Advanced Applications of CRISPR Screening: Target Discovery Strategies Behind a Nature Study

Advanced Applications of CRISPR Screening: Target Discovery Strategies Behind a Nature Study

Target identification remains one of the most critical and challenging steps in both basic and translational biomedical research. Discovering a novel therapeutic target can often lead to major breakthroughs and high-impact publications. Among the most powerful tools enabling systematic target discovery today is the CRISPR pooled screening platform, which allows high-throughput, genome-wide or pathway-specific functional interrogation.
In this article, we analyze a recent Nature publication that leveraged a targeted CRISPR knockout screen to dissect immune evasion mechanisms during early metastatic reactivation in lung cancer. Through this study, we will break down the design logic and execution of high-level CRISPR screening workflows and highlight key strategies for generating publishable, impactful results.

Nature Article
Scientific Background: DTCs and Immune Evasion in Early Metastasis
During metastasis, most disseminated tumor cells (DTCs) shed from the primary tumor are eliminated by the immune system. However, a small fraction survives, escapes immune surveillance, and enters a dormant state in distant organs. These dormant DTCs can downregulate MHC-I and NK cell-activating ligands, thereby avoiding detection by T cells and NK cells.
Eventually, some DTCs re-enter the cell cycle, transitioning into a phase of incipient metastasis.At this stage, the tumor cells exist in a delicate balance between immune visibility and immune evasion. Once this balance tips, DTCs rapidly proliferate, forming macrometastases.
This study aimed to uncover the immune regulators that suppress metastatic reactivation in the incipient phase, using a customized CRISPR knockout (CRISPRko) screen to identify key genes involved in immune control of early metastatic progression.
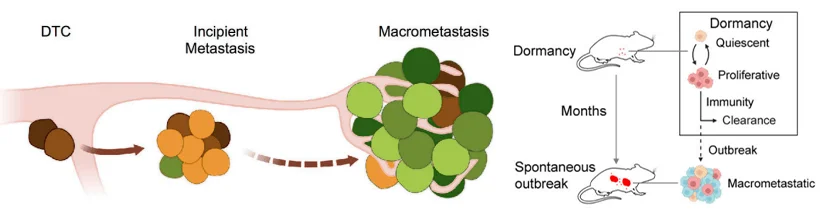
Figure 1. Tumor cell metastasis escape model [1,2]
CRISPR Screening Workflow: Design and Implementation
Step 1:Custom sgRNA Library Design: Focused and High-Fidelity
Key strategy: Build a compact, targeted sgRNA library tailored to immune regulation pathways.
- · Objective: Identify genes in DTCs that enable immune evasion and reactivation.
- · Scope: Identify genes in DTCs that enable immune evasion and reactivation.
-
· Threee primary gene categories:
- 1. MHC-I and NK ligand genes (targets of T and NK cell surveillance)
- 2. IFN-α/γ and complement system genes (identified via scRNA-seq and GSEA)
- 3. Upstream regulators like STING, RNA sensing, and LPS pathways
Each gene was targeted with 5 sgRNAs. The full library was divided into sub-libraries (~20 genes each) to ensure representative delivery and experimental control.
Step 2:Viral Transduction of Cancer Cell Lines: Ensuring Coverage and Representativeness
· Cell lines used: H2087-LCC and KPad1 (early-stage human lung adenocarcinoma)
· MOI: 0.3 to promote single-copy integration
· Coverage: >1000× to maintain sgRNA diversity
· Cell lines used: Puromycin (2.5 μg/mL) for 4 days post-transduction
Step 3:In Vivo Functional Screening: Modeling Immune Escape
Key strategy: Combine in vivo metastatic models with flow cytometry-based sorting of tumor cells.
- · Animal model: Immunodeficient mice to allow aggressive metastatic growth.
- · Injection method: Intracardiac or tail vein injection of transduced cells.
-
· Experimental design:
- 1. Control group: Dormant DTCs in early metastasis
- 2. Experimental group: Reactivated cells forming micrometastases
- · Screening method: In vivo imaging + FACS to isolate fluorescently labeled tumor cells from organs.
Each gene was targeted with 5 sgRNAs. The full library was divided into sub-libraries (~20 genes each) to ensure representative delivery and experimental control.
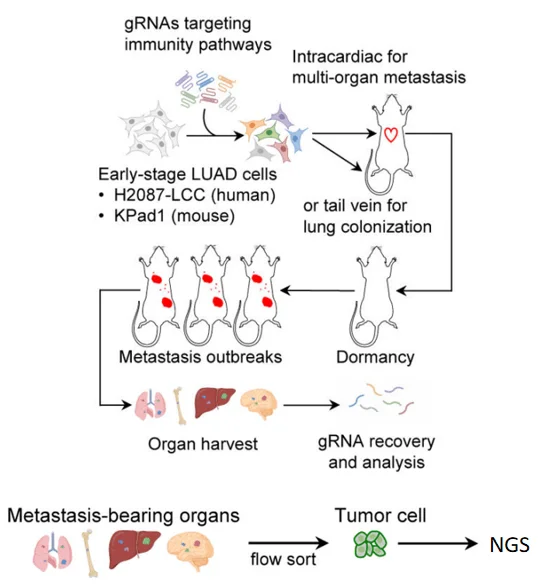
Figure 2. Flowchart of CRISPR library screening[1]
Step 4:NGS and Bioinformatics Analysis: Dual-Model Cross-Validation
· Sequencing: sgRNA abundance compared between early-stage and reactivated metastatic cells
· Key finding: sgRNAs targeting MHC-I and NK ligands were significantly enriched
· Validation: Both H2087-LCC and KPad1 models showed enrichment in STING pathway components
· Analysis pipeline: MAGeCK, edgeR, GSEA used for differential analysis and pathway enrichment
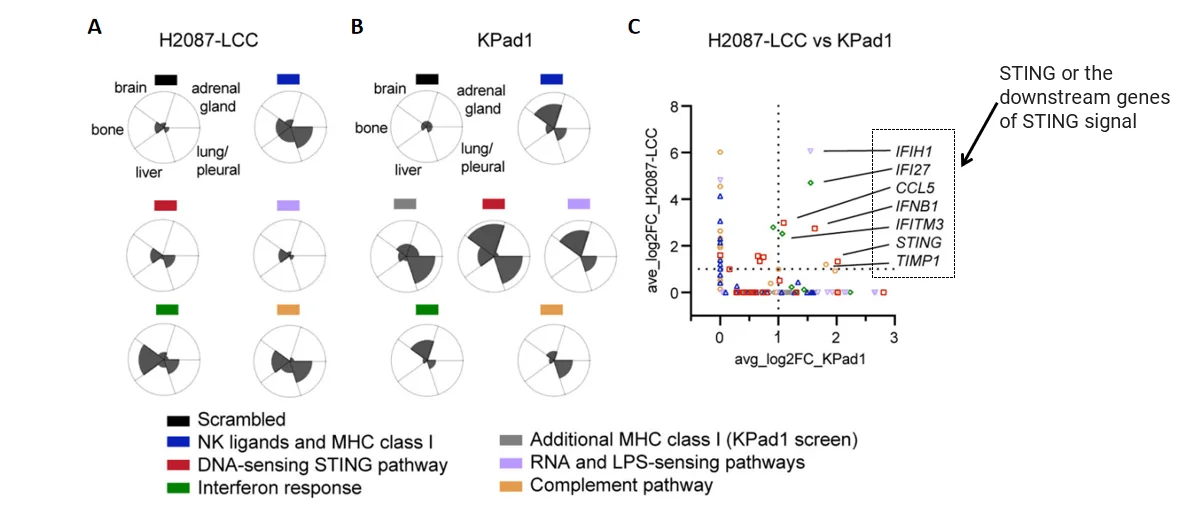
Figure 3. Screening targets by bioinformatics analysis[1]
In H2087-LCC cell infection group, sgRNA enrichment levels in different tissues;B. In Kpad1 cell infection group, sgRNA enrichment levels in different tissues; C. sgRNA enrichment analysis of the above two groups of cells
Step 5:Target Validation: STING as a Key Immunoregulatory Node
· Immunofluorescence: Confirmed increased STING expression in incipient metastatic cells
· Functional assays:
- 1. STING knockout → Accelerated metastatic reactivation
- 2. STING overexpression → Suppressed reactivation
This confirmed the critical role of the STING pathway in maintaining immune-mediated dormancy of DTCs.
![Target validation of STING[1]](/uploads/allimg/250619/target-validation-sting.webp)
Figure 4. Target validation of STING[1]
A&B.Comparison of STING signal intensity in two cell lines. Compared with the dormant cells (Ki67low), the STING signal of the cells at the incipient metastasis stage (Ki67high) was significantly increased; C. Knockout of Sting gene in KPad1 cells significantly increased the proliferation of tumor cells; D. Expression of Sting gene by Tet-On system significantly inhibited the proliferation of tumor cells; E. After the knockout of Sting gene, tumor cells in all organs significantly proliferated.
Summary: Four Key Elements of a High-Impact CRISPR Screen
| Step | Core Task | Highlight |
|---|---|---|
| Library Design | Targeted, custom sgRNA panels | Small, focused libraries increase specificity |
| Model Selection | Physiologically relevant systems | In vivo models reflect real tumor-immune dynamics |
| Data Analysis | Bioinformatic enrichment & filtering | Cross-validation across models boosts robustness |
| Target Validation | Functional gain/loss experiments | Completes the mechanistic discovery loop |
Know more about CRISPR screen>>>
Future Directions: Where is CRISPR Screening Headed?
· Single-cell CRISPR screens: Integrate with scRNA-seq for high-resolution dissection of gene function
· Multiplexed readouts: Combine with imaging, transcriptomics, or proteomics for multi-dimensional phenotyping
· In vivo pooled screens: Greater use of orthotopic and humanized models to capture complex microenvironments
· Pathway-aware library design: Expand to CRISPRa/i and dual-function libraries for modulating regulatory axes
· Drug-CRISPR co-screens: Combine small molecule exposure with genetic perturbation to identify synthetic lethal pairs
Stay connected with us for more case studies, technical resources, and tips for designing your own CRISPR screening strategy. Whether you are new to pooled libraries or exploring advanced applications, we are here to support your next discovery.
Need help designing a custom sgRNA library or CRISPR screening strategy? Contact our technical team today.
References
[1] Hu, J., Sánchez-Rivera, F.J., Wang, Z. et al. STING inhibits the reactivation of dormant metastasis in lung adenocarcinoma. Nature 616, 806–813 (2023).
[2] Laughney, A.M., Hu, J., Campbell, N.R. et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat Med 26, 259–269 (2020).


