Can CRISPR library be used to screen targets for non-proliferative phenotypes?

Can CRISPR library be used to screen targets for non-proliferative phenotypes?
As a useful tool for functional genomic target screening, CRISPR library has significant popularity nowadays. Recently, Ubigene received some messages for the same confusion about library screening: is it true that only phenotypes related to cell survival, proliferation and apoptosis can be screened by library? Otherwise, how can the target cells be enriched?
It is true that phenotypes with differences in cell proliferation and apoptosis are relatively easier to do CRISPR library screening, but it is also feasible to do screening on the phenotypes with non-proliferative differences. Through flow cytometry sorting, cells with antibody binding differences (membrane surface proteins) or fluorescence expression differences (reporter cell lines) can be sorted and enriched, and then the targets can be analyzed by NGS. Today, Ubigene will share an article published by Ryan J. Park et al. on Nature Genetics (IF=30.8): A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. This article used the CRISPR library to screen host factors related to HIV infection with the help of reporter cells and flow sorting[1].
HIV virus is a lentivirus that attacks the cells of the human immune system, which is a retrovirus. Its main target is the CD4+ T lymphocytes which play an important role in the human immune system. HIV virus causes the loss of immune function in the human body by destroying a large number of CD4+ T cells, thus causing AIDS, which seriously threatens human life and health.
Figure 1. HIV virus
At present, therapeutic drugs for HIV mainly directly target viral proteins, which are prone to acquired drug resistance mutations in clinical use. Compared with drugs that directly target viral proteins, drugs targeting the host protein required for HIV infection can improve the drug resistance barrier in anti-HIV treatment, which is a more ideal HIV treatment method. It has great theoretical significance and practical application value for elucidating the molecular mechanism of HIV breaking through the host cell defense system and developing new targets for HIV treatment.
CCR5 (C-C chemokine receptor type 5) is a co-receptor of the infection host CCR5-tropic HIV-1, which cooperates with CD4 to promote HIV virus to enter host cells by binding to HIV glycoprotein gp120.
In the early research of using RNAi to screen HIV therapeutic targets, nearly a thousand candidate host factors were obtained, but the results had the disadvantages of low overlap, incomplete gene silencing, and high off-target efficiency, etc. In this article, the authors used the method of genome-wide CRISPR knockout library screening to verify five host factors of CCR5 HIV strains in primary CD4+ T cells, which are the co-receptors CD4 and CCR5 which are necessary for HIV infection; TPST2 and SLC35B2, which play a role in the pathway of sulfating CCR5 on extracellular tyrosine residues, promote CCR5 to be recognized by HIV envelope; And the ALCAM gene, which mediates the transmission of HIV between cells. The knockout of the above genes does not affect the survival of cells but can promote cells to resist HIV infection, suggesting that HIV relies on a limited number of non-essential host proteins for replication. At the same time, the results also provide potential targets for HIV treatment and provide a new idea and way for the prevention and treatment of AIDS.
So, it can be seen that the library screening method has higher sensitivity and specificity than RNAi, and TPST2, SLC35B2, and ALCAM were not screened out in the early RNAi research.
The following is an introduction to the process of CRISPR library screening in this article.
1. Constructing a physiologically relevant CD4+ T cell line model
The authors selected CCRF-CEM cells, a CD4+ T-cell leukemia line to infect cas9 virus on CCRF-CEM cells stably expressing CCR5-hygro and HIV-1 LTR-GFP, and constructed a tool cell line (CXR-Cas9 cells) for screening HIV infection targets (Fig. 2). The cells highly expressed CCR5 and with low expression level of GFP before HIV-1 infection, but after HIV-1 infection, the expression of viral products increased the transcription of HIV-1 LTR promoter, resulting in high expression level of GFP, which allowed the detection of cell infection status by flow cytometry fluorescence sorting technology at the single-cell level.
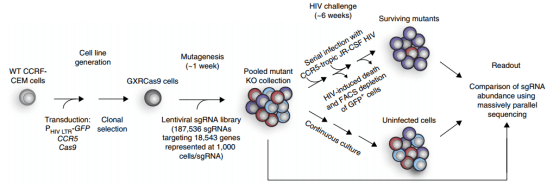
Figure 2. Genome-wide CRISPR screening of HIV host dependent factors
2. sgRNA library preparation and virus packaging
The library contains 187536 sgRNAs, targeting 18543 protein-coding genes (with an average of 10 sgRNAs per gene) and 1504 non-targeting control sgRNAs. The library was packaged into lentivirus then infected CXRCas9, then 24 hours upon infection, the successfully infected cells were screened by puromycin and were subsequently used for genome-wide screening.
After one week of infection with the library virus, these T cells lacking different receptors were infected with HIV strain JR-CSF at an infection MOI of 0.025, and GFP positive cells (~10-20%) could be detected after one week of infection. After another 2 weeks, the cells were infected with HIV strain JR-CSF under the same conditions and continued to be cultured for 10 days. It was found that there was no change in cell viability or GFP expression, indicating that the remaining cells were resistant to HIV infection after gene knockout (Fig. 3).
Compared with the parental cell lines, most of the surviving mutants lacked CD4 or CCR5 (Fig. 3), and the surviving cell population maintained high expression of CD4 and CCR5 on the cell surface, suggesting that the screening identified additional host factors for HIV infection. After another 3 weeks, viable GFP negative and positive cells were isolated by FACS, and the GFP negative cell population, the initial cell population collected before HIV infection, and the cell population that had not been infected with HIV virus for 6 weeks were sequenced to analyze the differences in sgRNA abundance. Finally, five genes with the largest changes in abundance were screened out (Fig. 4), Moreover, almost all gRNAs targeting two control genes, CXCR4 (encoding an HIV co-receptor not used by JR-CSF) and GTPase-encoding RAP2A, were not enriched (Fig. 5).
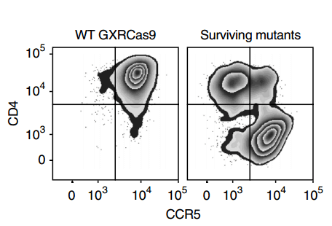
Figure 3. Flow cytometry analysis (representative of 3 independent experiments) quantified the expression of CD4 and CCR5 on the surface of wild-type GXRCas9 cells and GXRCas9 cells infected with a genome-wide sgRNA library and continuously infected with HIV-1 strain JR-CSF.
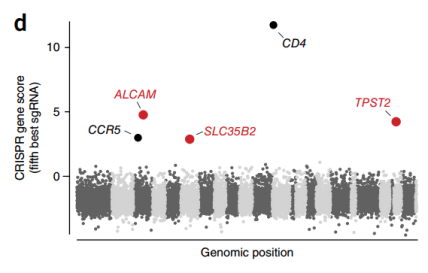
Figure 4. Fold change of the abundance of the fifth most enriched sgRNA of each gene after HIV infection by log2 transformation
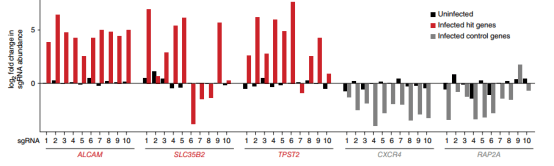
Figure 5. Single sgRNA enrichment of three candidate host dependent factors and two control genes
In order to explore how the deletion of TPST2, SLC35B2 and ALCAM genes can resist HIV infection, the authors constructed a clonal GXRCas9 cell line with these genes knocked out using CRISPR-Cas9 system and carried out a series of experiments to verify it. The results showed that TPST2 and SLC35B2 could promote the recognition of CCR5 by HIV envelope, while ALCAM molecule mediates the transmission of HIV between cells. This result provided potential targets for the treatment of HIV infection.
References
[1] Park R J, Wang T, Koundakjian D, et al. A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors[J]. Nature genetics, 2017, 49(2): 193-203.


