[Citation collection] Ubigene’s in-stock KO cell lines assist new progress in disease research
Ubigene focuses on the research and development and production of gene-editing cells, with nearly 3,000 square meters of gene editing technology platform, cell biology platform and antibody verification platform. At present, Ubigene has provided technical services and products to scientific research institutions and industrial customers in more than 30 countries and regions around the world.
With the exclusive CRISPR U™ patented technology, Ubigene has improved the editing efficiency of the traditional CRISPR/Cas9 to more than 10 times, and based on this, Ubigene has carried out the Red Cotton gene knockout project to build a gene-editing cell bank with more than 3,500 KO cells, and plans to expand to 5,000 cases by 2024.
Ubigene’s KO Cell Bank has 3500+ KO cells in stock, covering 26 signal pathways, 5 major drug targets, 27 disease types, etc., fast as 1-week delivery of homozygous clone, and Ubigene can also provide KO cell customization services! If you need services or products related to gene knockout, contact us for the solution suitable for your project >>>
KO cell lines are often used to study gene function, disease mechanism, drug screening, and so on. The following 9 articles related to the study of diseases have used the KO cell lines provided by Ubigene.
List
① SNORD17 knockout HepG2 cell line - Hepatocellular carcinoma IF=15.8
② TFEB Knockout L-02 cell line - Autophagy IF=13.30
③ Pik3r1 knockout RAW264.7 cell line - Osteoporosis IF=11.4
④ Knockout MC38 and CT26 - Colon cancer IF=10.75
⑤ PPIA knockout THP-1 - Inflammation IF=8.8
⑥ Astrin knockout Hela - Psoriasis IF=7.36
⑦ AM122A, PP5, DNA-PK knockout 293T - Cancer treatment IF=5.0
⑧ HIF-1α knockout RAW264.7 - Osteoporosis IF=4.63
⑨ TSC2 knockout NIH-3T3 - Tuberous sclerosis complex (TSC) IF=2.99
① SNORD17 knockout HepG2 cell line - Hepatocellular carcinoma
Published on Cell Death & Differentiation, IF=15.8
Title: Non-coding small nucleolar RNA SNORD17 promotes the progression of hepatocellular carcinoma through a positive feedback loop upon p53 inactivation
Published by researchers from Hepatic Surgery Center, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
Abstract: This research reveals the regulatory role of small nucleolus RNA SNORD17 and p53 signaling pathways in hepatocellular carcinoma, providing a new potential target for the treatment of hepatocellular carcinoma. Ubigene provided a key cell model for this study: SNORD17 knockout HepG2 cell line. In vitro and in vivo experiments showed that knockout of SNORD17 gene in HCC cell lines can significantly inhibit cell proliferation, clone formation and G1/S phase transformation[1].

Figure 1. The effect of SNORD17 mediated by p53 on HCC cell growth
② TFEB Knockout L-02 cell line - Autophagy
Published on Small, IF=13.30
Title: Silica Nanoparticles Trigger Chaperone HSPB8-Assisted Selective Autophagy via TFEB Activation in Hepatocytes
Published by researchers from Capital Medical University, Beijing, China
Abstract: Silica nanoparticles (SiNPs) are one of the most common inorganic nanomaterials, and autophagy is the predominant biological reaction to nanoparticles. This article used the TFEB knockout L-02 cell line built by Ubigene to verify that TFEB participates in SiNPs-induced autophagy. By comparing the transcriptomes of wild-type and TFEB KO cells using RNA sequencing, it was proved that SiNPs activate liver cells to trigger HSPB8-assisted selective autophagy through TFEB[2].

Figure 2. Diagram of CASA Mechanism induced by SiNPs
③ Pik3r1 knockout RAW264.7 cell line - Osteoporosis
Published on Redox Biology, IF=11.4
Title: GSTP1-mediated S-glutathionylation of Pik3r1 is a redox hub that inhibits osteoclastogenesis through regulating autophagic flux
Published by researchers from Orthopedics Research Institute of Zhejiang University, Hangzhou, China
Abstract: This article explores a new mechanism for GSTP1 affecting the osteoclast cell-related bone homeostasis through combing with a large number of in vivo and in vitro experiments, based on the Pik3r1 knockout RAW264.7 cell model built by Ubigene. It was the first time to interpret that the cell fate of osteoclasts is determined by S-glutathionylation via a redox-autophagy which is mediated by GSTP1[3].
④ TNFR2 knockout MC38 and CT26 - Colon cancer
Published on Int. J. Biol. Sci., IF=10.75
Title: TNFR2 deficiency impairs the growth of mouse colon cancer
Published by researchers from University of Macau, Macau, China
Abstract: This study used TNFR2 gene knockout MC38 and CT26 colon cancer cell lines provided by Ubigene, it was proved that TNFR2 deficiency can inhibit the AKT signaling pathway, providing further experimental evidence to support the view that blocking TNFR2 may represent a new strategy for cancer immunotherapy, and providing further evidence for developing TNFR2 antagonistic agents in the treatment of cancer[4].

Figure 3. TNFR2 gene knockout MC38 and CT26 colon cancer cell lines constructed by CRISPR/Cas9. (A) Flow cytometry analysis; (B) Real-time fluorescence quantitative analysis
⑤ PPIA knockout THP-1 - Inflammation
Published on Cell Reports, IF=8.8
Title: Delicate regulation of IL-1β-mediated inflammation by cyclophilin A
Published by researchers from Chinese Academy of Sciences, Beijing, China
Abstract: This article discussed the role and mechanism of CypA in the process of inflammation activation, resolution and tissue repair. The PPIA knockout THP-1 cell model constructed by Ubigene was used in the experiment. Through LPS, immunoblotting, phosphorylation and other experiments, it was proved that in the LPS-induced inflammation mouse model, CypA mainly regulates the expression of IL-1β to control its role in different stages of inflammation[5].
⑥ astrin knockout Hela - Psoriasis
Published on BMC Biology, IF=7.36
Title: CCHCR1-astrin interaction promotes centriole duplication through recruitment of CEP72
Published by researchers from Shenzhen University Health Science Centre, Shenzhen, China
Abstract: In order to verify the relationship between HCR protein and the interaction between the centrosome and mitosis-related proteins, the Hela cell lines with HCR and astrin knocked out respectively were used as the research material in this paper (the astrin knockout Hela was provided by Ubigene). The deletion of HCR or astrin significantly reduced the centrosome localization of CEP72 and subsequent MCPH proteins; The deletion of HCR also led to centriole duplication defects, mitotic errors, as well as multipolar spindle formation, genomic instability and DNA damage. This reveals that HCR could ensure effective centrosome replication and the stabilization of other centrosome-related functions by interacting with astrin (see Figure 4). This finding provides a deeper understanding of the molecular functions of HCR and helps to better explore the role of HCR in psoriasis and other diseases[6].
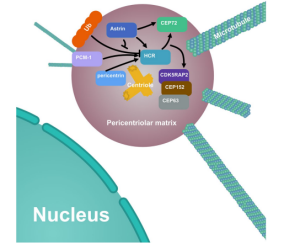
Figure 4. Localization and regulatory relationship of HCR and its related proteins on centrosomes
⑦ AM122A, PP5, DNA-PK knockout 293T - Cancer treatment
Published on Journal of Biological Chemistry, IF=5.0
Title: Phosphatases maintain low catalytic activity of SGK1: DNA damage resets the balance in favor of phosphorylation
Published by researchers from School of Medicine, Tsinghua University, Beijing, China (Cecilia Canessa research group)
Abstract: Using AM122A, PP5, DNA-PK knockout 293T cells provided by Ubigene, the researchers found from the experiment that the activity of SGK1 was inhibited by the action of S/T phosphatases PP5 and PP2A, which constantly dephosphorylated SGK1. The findings provide a molecular pathway that enables SGK1 to acquire phosphorylation and catalytic activity, and promote cell survival. This finding may have important implications for the development of novel cancer therapies targeting SGK1[7].
⑧ HIF-1α knockout RAW264.7 - Osteoporosis
Published on Bone, IF=4.63
Title: HIF-1α mediates osteoclast-induced disuse osteoporosis via cytoophidia in the femur of mice
Published by researchers from Stomatological Hospital and Dental School of Tongji University, Shanghai, China
Abstract: Using the HIF-1α knockout RAW264.7 constructed by Ubigene, this study found that knockout of this gene could improve osteoclastic bone resorption in the trabecular bone, revealing that HIF-1α gene plays a new role in supporting osteoclastogenesis and bone resorption, and provides direct evidence for its role in the pathogenesis of osteoporosis[8].
⑨ TSC2 knockout NIH-3T3 - Tuberous sclerosis complex (TSC)
Published on Cell Biochemistry and Biophysics, IF=2.99
Title: Construction of TSC2 knockout cell line using CRISPR/Cas9 system and demonstration of its effects on NIH-3T3 cells
Published by researchers from Peking Union Medical College Hospital, Beijing, China
Abstract: TSC2 gene plays an important role in the occurrence and development of Tuberous sclerosis complex (TSC). In this study, TSC2 knockout NIH-3T3 cell line constructed by Ubigene was used, and it was found that TSC2 knockout could enhance the proliferation and invasion of NIH-3T3 cells. In addition, KO cell line can simulate the pharmacological characteristics of TSC patients and can be used as an effective tool for further study of TSC[9].
Ubigene is committed to providing more gene-editing cell models for researchers worldwide, helping their research on disease mechanisms, disease diagnosis, drug research and development, gene therapy, and other research, and contributing to the development of human medicine!
References:
[1]Liang, J., Li, G., Liao, J. et al. Non-coding small nucleolar RNA SNORD17 promotes the progression of hepatocellular carcinoma through a positive feedback loop upon p53 inactivation. Cell Death Differ 29, 988–1003 (2022).
[2]Zhu Y, Zhang Y, Fan Z, et al. Silica Nanoparticles Trigger Chaperone HSPB8‐Assisted Selective Autophagy via TFEB Activation in Hepatocytes[J]. Small, 2023, 19(5): 2204310.
[3]Ji X, Hong J, Yang W, et al. GSTP1-mediated S-glutathionylation of Pik3r1 is a redox hub that inhibits osteoclastogenesis through regulating autophagic flux[J]. Redox Biology, 2023, 61: 102635.
[4]Li P, Yang Y, Yang X, et al. TNFR2 deficiency impairs the growth of mouse colon cancer[J]. International Journal of Biological Sciences, 2023, 19(4): 1024.
[5]Yang W, Bai X, Luan X, et al. Delicate regulation of IL-1β-mediated inflammation by cyclophilin A[J]. Cell reports, 2022, 38(11).
[6]Ying Z, Wang K, Wu J, et al. CCHCR1-astrin interaction promotes centriole duplication through recruitment of CEP72[J]. BMC biology, 2022, 20(1): 1-21.
[7]Gu W, Zheng H, Canessa C M. Phosphatases maintain low catalytic activity of SGK1: DNA damage resets the balance in favor of phosphorylation[J]. Journal of Biological Chemistry, 2023: 104941.
[8]Bie M, Tang Y, Xia Y, et al. HIF-1α mediates osteoclast-induced disuse osteoporosis via cytoophidia in the femur of mice[J]. Bone, 2023, 168: 116648.
[9]Wang X, Zhao Y, Wang Z, et al. Construction of TSC2 knockout cell line using CRISPR/Cas9 system and demonstration of its effects on NIH-3T3 cells[J]. Cell Biochemistry and Biophysics, 2022, 80(4): 681-687.
 Subscribe Us
Subscribe Us Gene Editing Services
Gene Editing Services
 EZ-editor™
EZ-editor™ Red Cotton Gene knockout Project
Red Cotton Gene knockout Project