"Knockout CHO Cells" (Chinese Hamster Ovary Cells) is a laboratory-cultured cell line technology derived from Chinese hamster ovary cells. Chinese hamsters are a popular laboratory mammal, partly due to their small size and low number of chromosomes, making them a good model for tissue culture and radiology research. CHO cells are commonly used mammalian cell models in biology, medicine and pharmaceutical research. In addition, they are often used for commercial purposes to produce therapeutic recombinant proteins. The growing demand for therapeutic proteins has led to the development of technologies that promote high quality and high productivity in the CHO expression system. The CHO culture process has been established, and the cost of using CHO cells to produce therapeutic proteins is equivalent to the cost of microbial culture.
Use CRISPR/Cas9 and fluorescence enrichment to generate triple knockout CHO cell lines in one step
It has previously been proven that CRISPR/Cas9 genome editing technology is an efficient tool for gene disruption in CHO cells. Stable Cell Lines In this study, the researchers further demonstrated the applicability and efficiency of CRISPR/Cas9 genome editing by simultaneously destroying FUT8, BAK and BAX in multiple settings in CHO cells. In order to isolate Cas9-expressing cells from the transfected cell bank, GFP was linked to Cas9 nuclease via 2A peptide. Using this method, the average indel frequency generated at the three genomic loci increased from 11% before enrichment to 68% after enrichment. Although there are a large number of genome editing events in the enriched cell pool, no obvious off-target effects are observed from off-target prediction and then deep sequencing. Single cell sorting was performed on the enriched multiplexed cells, and 97 clones were deeply sequenced, and it was found that there were 4 single gene, 23 double gene and 34 triple gene disrupted cell lines. Further characterization of selected potential tripartite knockout clones confirmed the removal of Bak and Bax proteins and disrupted fucosylation activity, as expected. Compared to wild-type CHO-S cells, the knock-out cell line showed improved resistance to apoptosis. In conclusion, multiplexing with CRISPR/Cas9 can further accelerate genome engineering work in CHO cells.
Knock out difficult-to-remove CHO host cell protein lipoprotein lipase to improve the stability of polysorbate in monoclonal antibody preparations
Although most host cell protein (HCP) impurities are effectively removed in a typical downstream purification process, a small portion of HCP is particularly challenging. Previous studies have identified HCPs that are challenging for a variety of reasons. Lipoprotein lipase (LPL) is a Chinese hamster ovary (CHO) HCP, which has the function of hydrolyzing the esters in triglycerides. It is one of the ten HCPs identified in previous studies, and they are easily retained in downstream processing. . Due to the structural similarity between polysorbate and triglyceride, LPL may degrade polysorbate 80 (PS-80) and polysorbate 20 (PS-20) in the final product formulation. In this work, it was found that recombinant LPL has enzymatic activity against PS-80 and PS-20 under the typical solution conditions of a series of mAb preparations. LPL knock-out CHO cells were created using CRISPR and TALEN technology. Compared with wild-type samples, the resulting cell culture harvest proved that polysorbate degradation was significantly reduced without significant impact on cell viability.
Optimized CRISPR/Cas9 strategy for homology‐directed multiple targeted integration of transgenes in CHO cells
Site-specific integration has emerged as a promising strategy for precise Chinese hamster ovary (CHO) cell line engineering and predictable cell line development (CLD). CRISPR/Cas9 with the homology-directed repair (HDR) pathway enables precise integration of transgenes into target genomic sites. However, inherent recalcitrance to HDR‐mediated targeted integration (TI) of transgenes results in low targeting efficiency, thus requiring a selection process to find a targeted integrant in CHO cells. Researcher explored several parameters that influence the targeting efficiency using a promoter ‐Trap‐based single‐ or double‐knock‐in (KI) monitoring system. A simple change in the donor template design by the addition of single‐guide RNA recognition sequences strongly increased KI efficiency (2.9–36.0 fold), depending on integration sites and cell culture mode, compared to conventional circular donor plasmids. Furthermore, sequential and simultaneous KI strategies enabled us to obtain populati ons with ~1–4% of double-KI cells without additional enrichment procedures. Thus, this simple optimized strategy not only allows efficient CRISPR/Cas9-mediated TI in CHO cells but also paves the way for the applicability of multiplexed KIs in one experimental step without the need for sequential and independent CHO–CLD procedures.
Ubigene has more than 400 types of primary cells, including epithelial cells, endothelial cells, smooth muscle cells and fibroblasts from different species, such as human, rat, and mouse. We can provide a validation report for each primary cell. Our primary cells have been widely used in many research institutes and pharmaceutical enterprises.
Reference
Grav L M, Lee J S, Gerling S, et al. One‐step generation of triple knockout CHO cell lines using CRISPR/Cas9 and fluorescent enrichment[J]. Biotechnology Journal, 2015, 10(9): 1446-1456.
Chiu J, Valente K N, Levy N E, et al. Knockout of a difficult-to-remove CHO host cell protein, lipoprotein lipase, for improved polysorbate stability in monoclonal antibody formulations[J]. Biotechnology and Bioengineering, 2017, 114(5): 1006-1015.
Shin S W, Lee J S. Optimized CRISPR/Cas9 Strategy for Homology‐Directed Multiple Targeted Integration of Transgenes in CHO Cells[J]. Biotechnology and Bioengineering, 2020.
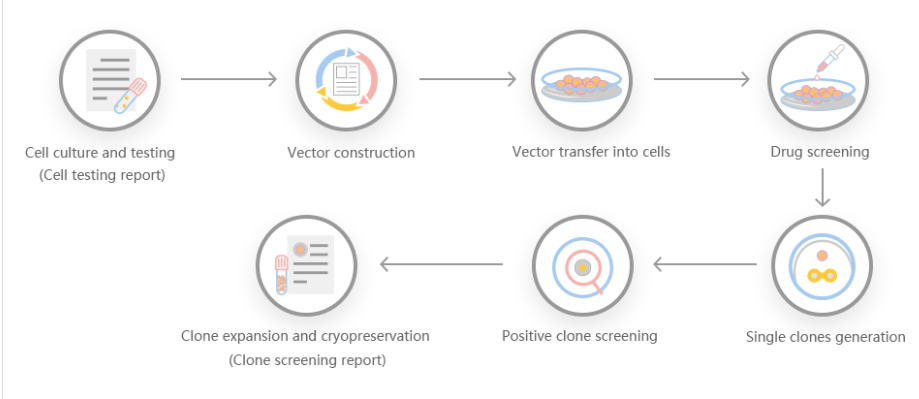
The efficiency of gene knock-out and cleavage can not only give people the ability to generate protein radical profiles and establish regulatory records, but also has many advantages, making it a particularly attractive recombinant protein expression system. First, it is carboxylated on glutamic acid and sulfated on tyrosine. Second, the operation is simple, and the recombinant protein can be quickly produced through transient gene expression. Third, it can be used for stable recombinant protein production. Some researchers used gene cell knockout and cutting efficiency systems to generate gene-edited cell lines, targeted sequencing of GLUL genomic loci, produced stable EPO cell lines, and discovered the mechanism of stable expression of recombinant erythropoietin in humans .
According to customer needs, Yuanjing Biotechnology designs a stable gene transfer knockout program based on the target gene
Scheme 1: Small-segment gene knockout program, gRNA is set in the introns at both ends of exon 2, and the number of bases encoded by the knockout exon is not 3 times, and the knockout can cause frameshift.
Scheme 2: Frameshift gene knockout scheme, gRNA is set on the exon, the number of missing bases is not 3 times, and frameshift mutation can occur after knockout.
Scheme 3: Large-segment gene knockout scheme, knock out the coding sequence of the entire gene to achieve the effect of large-segment knockout.
Ubigene Biosciences is co-founded by biological academics and elites from China, the United States, and France. We are located in Guangzhou Science City, which serves as a global center for high technology and innovation. Ubigene Biosciences has 1000㎡ office areas and laboratories, involving genome editing, cell biology technology, and zebrafish research. We provide products and services for plasmids, viruses, cells, and zebrafish. We aim to provide customers with better gene-editing tools for cell or animal research.
We developed CRISPR-U™ and CRISPR-B™(based on CRISPR/Cas9 technology) which is more efficient than general CRISPR/Cas9 in double-strand breaking, CRISPR-U™ and CRISPR-B™ can greatly improve the efficiency of homologous recombination, easily achieve knockout (KO), point mutation (PM) and knockin (KI) in vitro and in vivo.
Genome Editing Platform
——Focusing on the Application of CRISPR-U™ and CRISPR-B™ Gene Editing Technology
1. Provides various types of gene-editing vectors for different species.
2. Provides different virus packaging services, including lentiviruses, adenoviruses and adeno-associated viruses.3. Provides high-quality services for gene knockout, point mutation and knockin cell lines.
Cell Biology Platform
——Focusing on primary cell
1. Provides over 400 types of primary cells.
2. Provides culture strategies and related products for different cell types.3. Provides cell biology-related services such as cell isolation, extraction and validation.
 Subscribe Us
Subscribe Us Gene Editing Services
Gene Editing Services
 EZ-editor™
EZ-editor™ Red Cotton Gene knockout Project
Red Cotton Gene knockout Project