Location:Home > Application > Knockout cell line | circRNA' functio

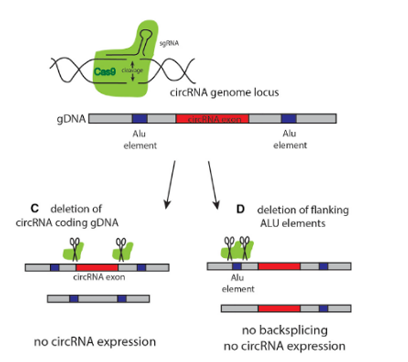
More and more researchers have given up using this strategy to knockout the circRNA because of its great influence on the parent gene. The ideal method is to knockout the loop forming elements (Alu) in the flanking intron of the exon, so as to destroy the circRNA loop forming without affecting the expression of the coding gene. However, it is difficult for many researchers to master the logic of designing the targeting strategy.
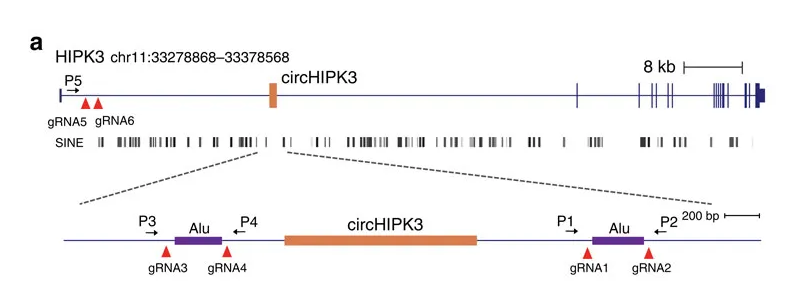
Taking circ-HIPK3 as an example, circ-HIPK3 is a kind of circRNA rich in human cells, which can combine with a variety of miRNAs as a regulator of cell growth and affect the formation of tumors. In order to verify how circ-HIPK3 forms into circle, it is necessary to find the loop forming elements in flanking intron. A pair of sgRNA is designed for the two Alu elements predicted at upstream and downstream respectively. The predicted loop forming elements are knockout by CRISPR/Cas9 system to detect whether the expression of circRNA changes. After PCR and RT-qPCR verification, it was found that the expression of circ-HIPK3 was significantly down-regulated after knockout of downstream loop forming elements, while the expression of circ-HIPK3 was not decreased but increased after knockout of upstream loop forming elements. It was speculated that there were too many loop forming elements in the upstream and the prediction was not accurate. In order to further verify the RNA circulation driven by other elements, the large fragment of intron in the upstream of the element was knockout by co-injection of gRNA3 or gRNA4 with gRNA5 or gRNA6. RT-qPCR results showed that the expression of circ-HIPK3 decreased, indicating that other loop forming elements of circ-HIPK3 exist.
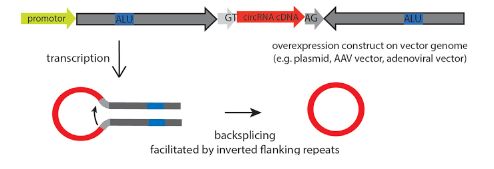
Overexpression of circRNA is not easy because of its low efficiency of loop forming, and it is easy to mismatch. By optimizing the binding sites of RBP, such as Alu elements and QKI, the circRNA can be formed accurately and efficiently. After overexpression, it is necessary to detect successful loop-forming and linear mRNA expression. In order to study the loop-forming efficiency of a new circRNA expression system, the mouse circrtn4 gene was selected to express in a variety of cell lines (including HeLa, N2a, HEK293). According to the RT-qPCR results in different cell lines, the efficiency of the new vector system pCircRNA-DMo-Rtn4 is much higher than that of the common vector system (pCircRNA-BE-Rtn4) in these cell lines.
CRISPR-U™ high-efficiency gene editing system:
Ubigene focuses on genome editing, CRISPR-U™ is a gene-editing technology developed by Ubigene, which is more efficient than common CRISPR/Cas9 technologies in gene targeting. The services related to circRNA editing introduced in this paper can be provided by Ubigene, including knockout, interference, overexpression and circRNA expression testing.

More and more researchers have given up using this strategy to knockout the circRNA because of its great influence on the parent gene. The ideal method is to knockout the loop forming elements (Alu) in the flanking intron of the exon, so as to destroy the circRNA loop forming without affecting the expression of the coding gene. However, it is difficult for many researchers to master the logic of designing the targeting strategy.

Taking circ-HIPK3 as an example, circ-HIPK3 is a kind of circRNA rich in human cells, which can combine with a variety of miRNAs as a regulator of cell growth and affect the formation of tumors. In order to verify how circ-HIPK3 forms into circle, it is necessary to find the loop forming elements in flanking intron. A pair of sgRNA is designed for the two Alu elements predicted at upstream and downstream respectively. The predicted loop forming elements are knockout by CRISPR/Cas9 system to detect whether the expression of circRNA changes. After PCR and RT-qPCR verification, it was found that the expression of circ-HIPK3 was significantly down-regulated after knockout of downstream loop forming elements, while the expression of circ-HIPK3 was not decreased but increased after knockout of upstream loop forming elements. It was speculated that there were too many loop forming elements in the upstream and the prediction was not accurate. In order to further verify the RNA circulation driven by other elements, the large fragment of intron in the upstream of the element was knockout by co-injection of gRNA3 or gRNA4 with gRNA5 or gRNA6. RT-qPCR results showed that the expression of circ-HIPK3 decreased, indicating that other loop forming elements of circ-HIPK3 exist.

Overexpression of circRNA is not easy because of its low efficiency of loop forming, and it is easy to mismatch. By optimizing the binding sites of RBP, such as Alu elements and QKI, the circRNA can be formed accurately and efficiently. After overexpression, it is necessary to detect successful loop-forming and linear mRNA expression. In order to study the loop-forming efficiency of a new circRNA expression system, the mouse circrtn4 gene was selected to express in a variety of cell lines (including HeLa, N2a, HEK293). According to the RT-qPCR results in different cell lines, the efficiency of the new vector system pCircRNA-DMo-Rtn4 is much higher than that of the common vector system (pCircRNA-BE-Rtn4) in these cell lines.

CRISPR-U™ high-efficiency gene editing system:
Ubigene focuses on genome editing, CRISPR-U™ is a gene-editing technology developed by Ubigene, which is more efficient than common CRISPR/Cas9 technologies in gene targeting. The services related to circRNA editing introduced in this paper can be provided by Ubigene, including knockout, interference, overexpression and circRNA expression testing.