Expert Insights | Understanding Common Methods for Point Mutation Research

Since April 1996, when the Human Gene Mutation Database (HGMD) first published 600 genes with approximately 10,000 gene mutations, it has continued to expand with new mutation information year by year. As of the writing of this article (July 24, 2024), 291,339 distinct gene mutations have been recorded, all of which are considered to be closely related to human genetic diseases. Among these, Missense/nonsense mutations are the most prominent, accounting for 58%, making them the most common pathogenic type.
To study the relationship between point mutations and diseases, screen drugs for treating point mutation diseases, or develop new therapies targeting point mutation diseases, it is usually necessary to first construct point mutation models. This article introduces three common methods for point mutation research in in vitro cell experiments, ranging from beginner to intermediate to advanced levels. Let's grab the notebooks and learn together with Ubigene.
Beginner Level: Overexpression of Mutant Transcripts
Method Description
In wild-type cells (without any gene editing), expression vectors containing mutant genes are introduced through transient or stable transfection methods, allowing the mutant genes to be expressed in the cells. This is typically used for preliminary screening, and the screened mutations require further experimental validation.
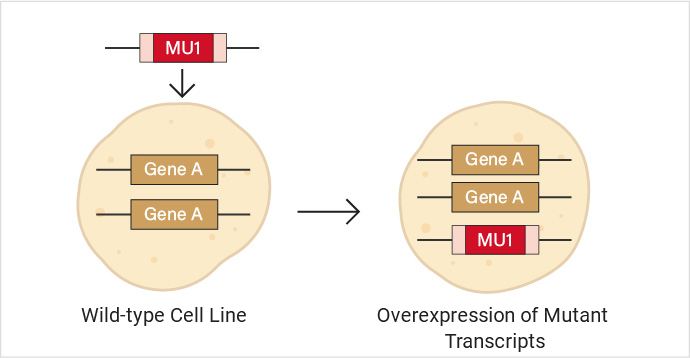
Diagram of overexpression of mutant transcripts
Advantages
1. Simple operation: No need for complex gene editing such as gene knockout; only requires constructing expression vectors and performing transfection or transduction, with the lowest operational difficulty.
2. Shortest turnaround time: Can quickly obtain cell lines expressing mutant genes; if using transient transfection methods, preliminary screening results can be obtained in as fast as a few weeks.
3. High expression levels: Generally achieves high levels of mutant gene expression, facilitating detection and analysis of gain-of-function effects under high expression conditions.
Disadvantages
1. Non-physiological expression levels: High levels of exogenous gene expression may lead to non-physiological phenotypes, affecting result interpretation.
2. Endogenous gene interference, uncertain phenotype: Normal expression of endogenous genes may interfere with the expression of exogenous mutant genes, making it difficult to observe mutant phenotypes.
3. Not applicable to non-coding regions: This method has limitations for studying point mutations in non-coding regions such as promoters and introns.
Case Study
Horsch et al. overexpressed wild-type KRAS and its mutants at positions 12 and 13 (KRASOE, KRASG12V, KRASG13D) in mouse fibroblast NIH3T3 cells, simultaneously measuring Kras mRNA levels, Ras protein, and activated Ras protein levels. Additionally, they used whole-genome expression profiling to study differences between the three transfected cell groups. Finally, by identifying and characterizing genes differentially regulated by Kras overexpression and different mutant forms, they provided new insights into understanding how this oncogene regulates cell transformation and tumor development[1].
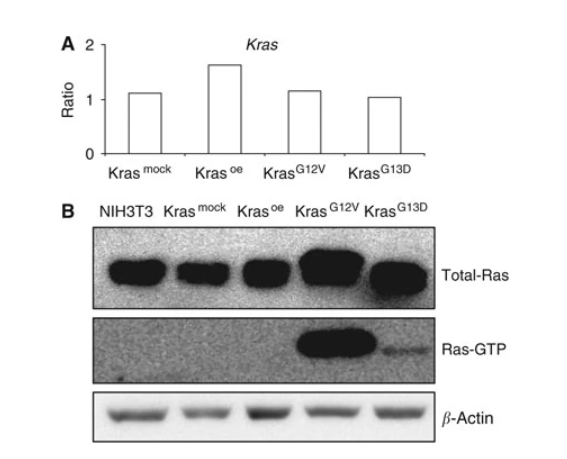
Figure 1. Detection after overexpression of KRAS wild-type and different mutants
Intermediate Level: KO+Overexpression of Mutant Transcripts
Method Description
First, the endogenous gene is knocked out using gene editing techniques such as CRISPR/Cas9, then mutant genes are introduced for overexpression in the knockout background. This approach can be conducted simultaneously with wild-type transcript overexpression, equivalent to performing a rescue experiment to verify gene function. This method is suitable for studying multiple point mutations with limited budget or when multiple mutation sites are far apart and require multiple point mutations, serving as an alternative option. It's important to note that this method requires preventing gRNA from cutting the overexpressed transcripts. Typically, gRNAs can be designed in different introns to knock out entire exons, or the RNP method can be used for endogenous gene KO to avoid this issue.
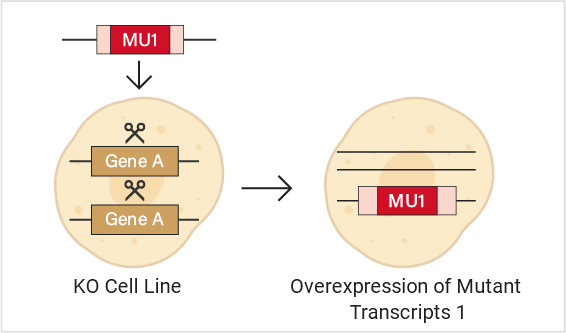
Diagram of KO+overexpression of mutant transcripts
Advantages
1. No endogenous gene interference: After knocking out the endogenous gene, exogenous mutant genes are not subject to competitive interference from endogenous genes.
2. Controllable expression: For studies requiring different expression level ratios of wild-type and mutant types, the amount of expression vector can be adjusted for control.
3. Lower difficulty: KO efficiency is higher than endogenous point mutations, with moderate technical difficulty.
4. Reduced cost for multiple site mutations: For studying multiple mutation sites, first performing KO and then overexpressing different mutant transcripts can reduce costs.
Disadvantages
1. Longer turnaround time: Requires two steps of gene editing, resulting in higher time costs.
2. Non-physiological expression levels: The expression level of mutant genes differs from natural physiological conditions, potentially affecting result interpretation.
3. Not applicable to non-coding regions: This method has limitations for studying point mutations in non-coding regions such as promoters and introns.
Case Study
Chen et al. performed high-depth whole-genome sequencing (average 120x) on tumor tissues from 494 hepatocellular carcinoma patients from different regions of China, conducting in-depth analysis of driver genes in coding and non-coding regions, mutation signatures, copy number variations, clustered mutation events, extrachromosomal circular DNA, and mutation evolution patterns. Additionally, they selected three newly identified potential driver events for detailed functional validation. Among these, the in situ point mutation experiment for the FGA gene failed. The authors adjusted their approach by first constructing FGA gene knockout HepG2 cells, then using lentiviral packaging and infection to express different point mutant transcripts for functional studies[2].
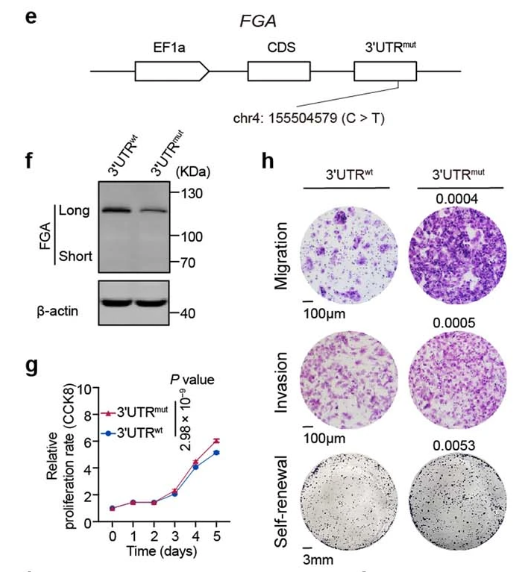
Figure 2. Expression levels and genotypes of FGA gene 3'UTR wild-type and mutant
Advanced Level: Endogenous Gene Site-Directed Mutagenesis
Method Description
Using gene editing techniques such as CRISPR/Cas9, gRNA, Cas9 protein, and a donor carrying the point mutation and homologous sequences are introduced into cells together. Through homologous recombination repair, specific site mutations are introduced without altering other parts of the gene. This method is closest to the physiological state and is the best choice for constructing disease models.
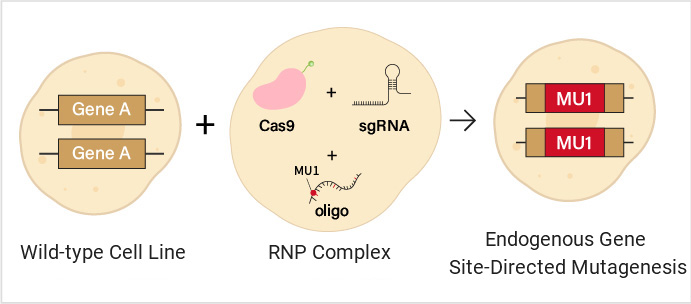
Diagram of endogenous gene site-directed mutagenesis
Advantages
1. High physiological relevance: Mutant genes work in their natural expression background, closer to physiological conditions, more suitable for studying pathological mechanisms.
2. Single-step operation: Only requires one gene editing process, completed in one experiment.
3. High safety: Generally uses the RNP method, with low off-target risk and no random integration of exogenous genes, resulting in high reliability of results.
4. Applicable to non-coding regions: Suitable for studying point mutations in non-coding regions such as promoters and introns.
Disadvantages
1. Higher technical difficulty: Homologous recombination repair efficiency is relatively low, requiring highly efficient gRNA and Cas9 protein. The construction process requires screening a large number of single-cell clones, demanding high experience.
2. High cost for multiple site mutations: When studying multiple mutation sites, if mutating multiple distant sites simultaneously or mutating multiple different sites separately, multiple point mutation gene editing experiments are required, thus increasing the cost of this method.
Case Study
Thyroid cancer is a common endocrine malignancy, with PTC being the most common type. Common driver gene alterations include BRAF mutations, RAS mutations, RET/PTC rearrangements, and TERT gene promoter mutations. The coexistence of BRAF/RAS mutations and TERT promoter mutations is associated with poor clinical outcomes in PTC, but the clinical behavior and carcinogenic nature of RET/PTC rearrangements in PTC remain controversial. Zhang et al. used CRISPR/Cas9 technology to repair TPC-1 cells containing the TERT C228T mutation to wild-type, finding that TERT gene expression was significantly downregulated, and RET/PTC inhibitors had no effect on the repaired wild-type TPC-1, demonstrating the dependence of RET/PTC action on TERT promoter mutations[3].
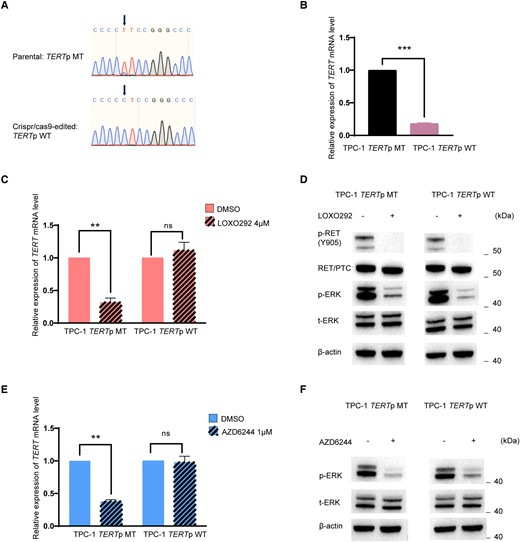
Figure 3. RET/PTC regulation depends on TERT promoter mutation
In conclusion, endogenous gene site-directed mutagenesis is the preferred method for point mutation research. However, if cost limitations or the difficulty of directly performing multiple point mutations are considered, an alternative approach can be chosen: KO+overexpression of mutant transcripts, but care must be taken to avoid gRNA cutting issues. In the early stages of a project, if rapid screening of multiple mutation sites is desired to preliminarily understand the functional phenotypes of point mutations, transient overexpression of these mutant genes in wild-type cells can be used to observe initial effects.
References
[1] Horsch, M., et al. "Overexpressed vs mutated Kras in murine fibroblasts: a molecular phenotyping study." British journal of cancer 100.4 (2009): 656-662.
[2] Chen, Lei, et al. "Deep whole-genome analysis of 494 hepatocellular carcinomas." Nature 627.8004 (2024): 586-593.
[3] Zhang, Wei, et al. "Coexisting RET/PTC and TERT Promoter Mutation Predict Poor Prognosis but Effective RET and MEK Targets in Thyroid Cancer." The Journal of Clinical Endocrinology & Metabolism (2024): dgae327.
 Subscribe Us
Subscribe Us Gene Editing Services
Gene Editing Services
 EZ-editor™
EZ-editor™ Red Cotton Gene knockout Project
Red Cotton Gene knockout Project