

Location:Home > Application > Ubigene KO Cells Help Reveal PCSK9 Protein Interactions And Regulation Of Lipid Metabolism
Ubigene KO Cells Help Reveal PCSK9 Protein Interactions And Regulation Of Lipid Metabolism

Background
PCSK9 (Proprotein convertase subtilisin/kexin type 9) is a protein associated with familial hypercholesterolemia. Its C-terminal M2 repeat sequence is crucial for its extracellular function, possibly interacting with an unknown "protein X". Recent studies show that the M2 repeat sequence can bind to the R-x-E motif in MHC-I class proteins (such as HLA-C), leading to their lysosomal degradation, suggesting a potential new role for PCSK9 in the immune system. Additionally, the mechanism of HFE's role in lipid metabolism remains unclear, and HLA-C is thought to potentially be involved in PCSK9-mediated LDLR degradation.
Abstract
Recently, the Nabil G Seidah research group published a research paper titled "Insights into PCSK9-LDLR Regulation and Trafficking via the Differential Functions of MHC-I Proteins HFE and HLA-C" in Cells. This study compared the effects of MHC-I-like proteins on PCSK9's extracellular function, revealing that HFE is a new target of PCSK9 that inhibits its activity on LDLR, while HLA-C enhances its function. The study used HLA-C gene knockout HepG2 cells constructed by Ubigene to investigate the regulation and trafficking mechanisms of PCSK9 and LDLR (low-density lipoprotein receptor), as well as the different functions of MHC-I proteins HFE and HLA-C in this process.
Results & Discussion
This study explored the interactions between HFE, HLA-C, and PCSK9 and their effects on LDLR function. The authors added HFE, HLA-C, and their chaperone β2M to PCSK9 gene knockout HepG2 cells and HLA-C gene knockout HepG2 cells (constructed by Ubigene), discovering that HFE can act as a negative regulator of PCSK9 on LDLR, inhibiting its degradation of LDLR. Meanwhile, extracellular PCSK9 enhanced the degradation of HFE and LDLR in acidic compartments. Further experiments suggested that the dissociation of HFE from TfR1 might increase the availability of HFE on the cell surface, thus inhibiting PCSK9's function on LDLR to a greater extent.
The study also found that HLA-C significantly enhanced PCSK9's function, confirming that HLA-C is a key factor in PCSK9-induced LDLR degradation. In CHO-K1 cells, overexpression of HLA-C significantly enhanced the degradation of LDLR by exogenous PCSK9. In HepG2 CRISPR HLA-C KO cells, the absence of HLA-C weakened PCSK9's effect on LDLR degradation, while overexpression of HLA-C restored PCSK9's function. These results suggest that HLA-C and HFE may regulate PCSK9's function through different mechanisms, among HFE mainly through inhibiting PCSK9's action on LDLR, while HLA-C through enhancing PCSK9's function to promote LDLR degradation.
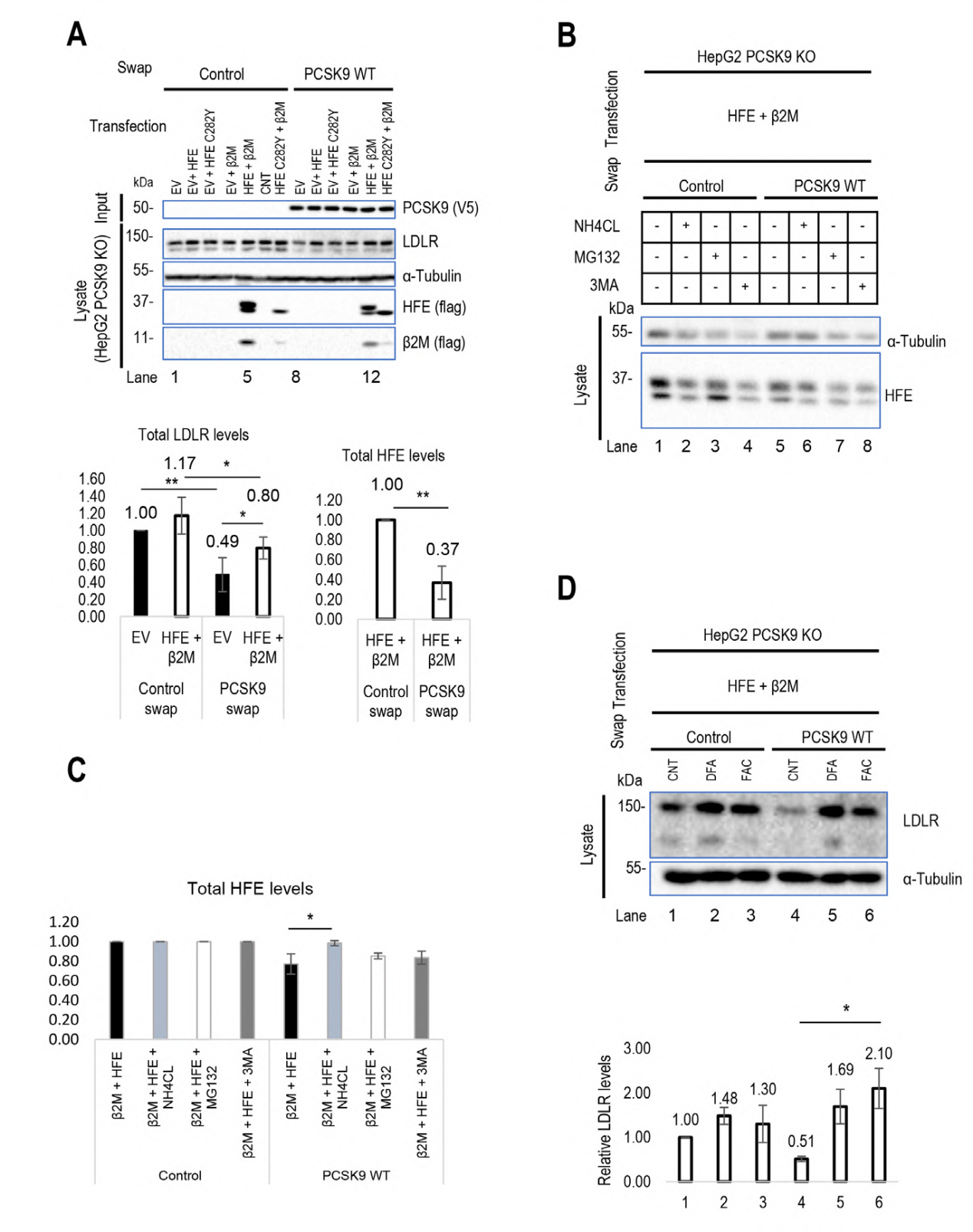
Figure 1 Regulatory effects of HLA-C and HFE on PCSK9
The authors used 3D modeling to reveal the importance of HLA-C and PCSK9 interaction and predicted their binding sites at the molecular level. The study found that the R68-G-E70 motif on HLA-C interacts with the M2 domain of PCSK9, while HFE, due to structural similarity, may bind to PCSK9's M2 domain through its R67-V-E69 motif. The interaction between HFE and PCSK9 was further validated using the GRAMM-X Web server for modeling. In experiments, the importance of these sites in the interaction between HFE and PCSK9 was confirmed by mutating or deleting key binding sites on PCSK9 and HFE. Furthermore, the study found that although HFE and HLA-C interact with the same residues of PCSK9, they may encounter PCSK9 through different transport pathways, thus affecting PCSK9's activity on LDLR. These findings provide new perspectives for understanding the interactions between HLA-C, HFE, and PCSK9 and their roles in lipid metabolism.
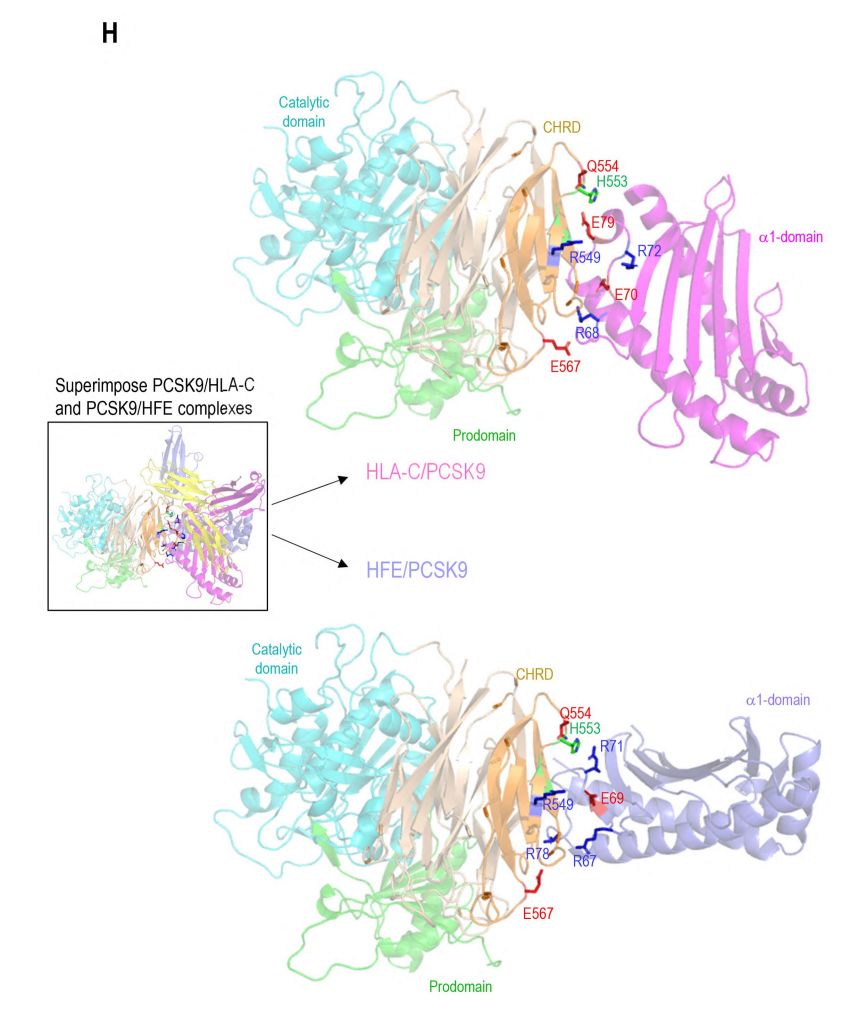
Figure 2 Direct interaction of PCSK9 with HLA-C compared to HFE
To investigate the internalization pathways of LDL receptor, HLA-C, and HFE and their interactions with PCSK9, the authors used methods to knock down caveolin-1 (siCav1) or clathrin heavy chain (siCHC). The results showed that HLA-C degradation mainly occured through clathrin-coated vesicles, while HFE internalization may depend on the combined action of caveolin and clathrin. The experiments also found that the presence of HFE seemed to inhibit PCSK9's function on LDLR, but further testing using the CircuLex Human PCSK9 Functional Assay Kit showed that the binding affinity between PCSK9 and LDLR remained unchanged regardless of HFE's presence. This suggests that HFE does not compete with LDLR for PCSK9 binding, but rather relies on LDLR's presence to be degraded by PCSK9. Additionally, the lack of LDLR inhibits PCSK9's function on HFE, while HLA-C degradation is not dependent on LDLR. The study also found that the cytoplasmic tail of LDLR is not necessary for PCSK9 to enhance LDLR degradation, while HLA-C plays a key role in sorting the PCSK9-LDLR complex to degradation compartments. These findings help to deepen the understanding of the regulatory mechanisms of LDLR and related proteins in lipid metabolism.
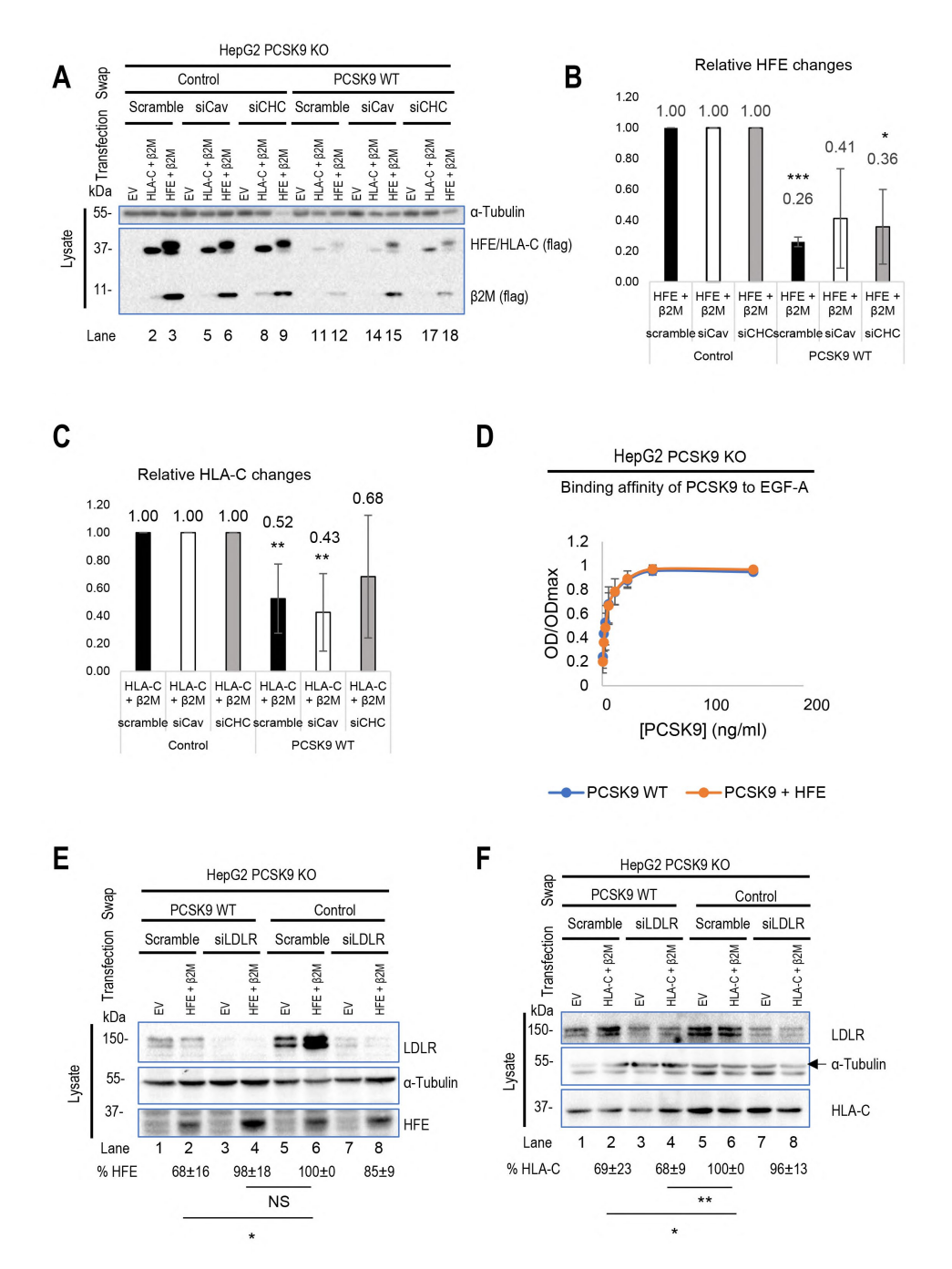
Figure 3 Transport differences between HLA-C and HFE
The authors also used 3D modeling technology to analyze in depth the interactions between PCSK9 and HLA-C and HFE. The results showed that both HLA-C and HFE bind to specific residues E567 and E549 in the CHRD region of PCSK9 through their α1 chains. Although the binding modes of both to PCSK9 are similar, there are subtle differences in the orientation of the α1 chains. At the same time, the N-terminal peptide of PCSK9 has the best interaction with the α1 chain of HLA-C, which was confirmed by Alphafold prediction and modeling. The interaction between the unstructured N-terminal residues of PCSK9 and the peptide-binding pocket of HLA-C shows high confidence, and this binding may be influenced by the structure of PCSK9's N-terminal peptide. Finally, the authors also constructed a complete PCSK9/HLA-C α1 chain/β2-microglobulin ternary model, supporting the view that PCSK9/HLA-C interact through CHRD and N-terminal peptides. This interaction involves residues R68 and E70 on the α1 chain of HLA-C, and residues E567 and E549 on the M2 module of CHRD. In addition, the study also evaluated the similarity of HLA-C antigen pockets to other MHC-I molecules, finding that positively charged residues are generally present in HLA-C antigen pockets, which may support the binding of acidic peptides.
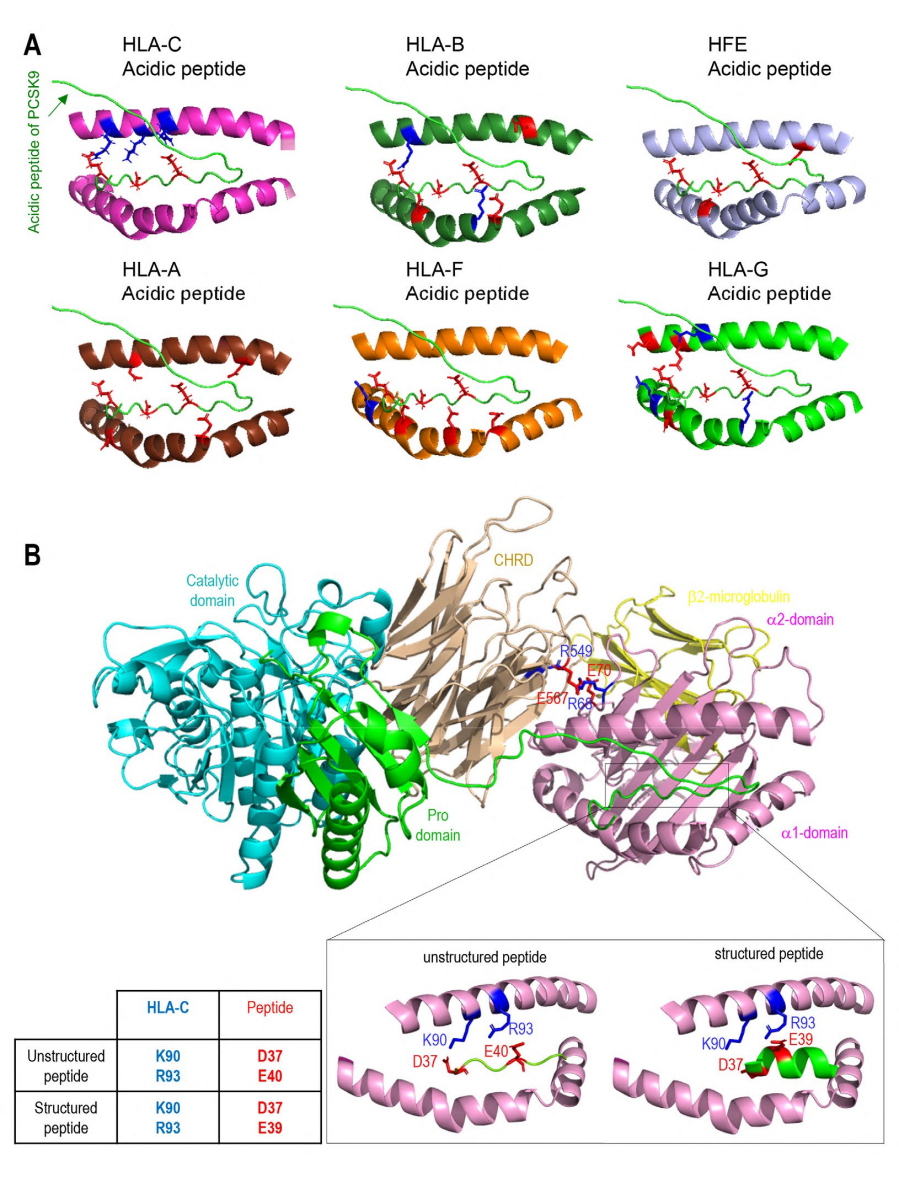
Figure 4 Modeling of interactions between PCSK9 N-terminus and HLA members
Conclusion
This study focuses on the interactions between PCSK9 and LDLR during lysosomal degradation, emphasizing the important roles of PCSK9's M2 domain and HLA-C molecule in this mechanism. It also reveals that HFE and HLA-C exhibit opposite effects in regulating PCSK9 function, and they adopt different endocytosis pathways when interacting with PCSK9. These key findings not only enrich our understanding of protein interactions in cellular transport processes but also provide new directions and perspectives for future research.
Reference
Nabil G. Seidah, et al. “Insights into PCSK9-LDLR Regulation and Trafficking via the Differential Functions of MHC-I Proteins HFE and HLA-C” Cells 5.1 (2024): 13(10): 857

Ubigene KO Cells Help Reveal PCSK9 Protein Interactions And Regulation Of Lipid Metabolism

Background
PCSK9 (Proprotein convertase subtilisin/kexin type 9) is a protein associated with familial hypercholesterolemia. Its C-terminal M2 repeat sequence is crucial for its extracellular function, possibly interacting with an unknown "protein X". Recent studies show that the M2 repeat sequence can bind to the R-x-E motif in MHC-I class proteins (such as HLA-C), leading to their lysosomal degradation, suggesting a potential new role for PCSK9 in the immune system. Additionally, the mechanism of HFE's role in lipid metabolism remains unclear, and HLA-C is thought to potentially be involved in PCSK9-mediated LDLR degradation.
Abstract
Recently, the Nabil G Seidah research group published a research paper titled "Insights into PCSK9-LDLR Regulation and Trafficking via the Differential Functions of MHC-I Proteins HFE and HLA-C" in Cells. This study compared the effects of MHC-I-like proteins on PCSK9's extracellular function, revealing that HFE is a new target of PCSK9 that inhibits its activity on LDLR, while HLA-C enhances its function. The study used HLA-C gene knockout HepG2 cells constructed by Ubigene to investigate the regulation and trafficking mechanisms of PCSK9 and LDLR (low-density lipoprotein receptor), as well as the different functions of MHC-I proteins HFE and HLA-C in this process.
Results & Discussion
This study explored the interactions between HFE, HLA-C, and PCSK9 and their effects on LDLR function. The authors added HFE, HLA-C, and their chaperone β2M to PCSK9 gene knockout HepG2 cells and HLA-C gene knockout HepG2 cells (constructed by Ubigene), discovering that HFE can act as a negative regulator of PCSK9 on LDLR, inhibiting its degradation of LDLR. Meanwhile, extracellular PCSK9 enhanced the degradation of HFE and LDLR in acidic compartments. Further experiments suggested that the dissociation of HFE from TfR1 might increase the availability of HFE on the cell surface, thus inhibiting PCSK9's function on LDLR to a greater extent.
The study also found that HLA-C significantly enhanced PCSK9's function, confirming that HLA-C is a key factor in PCSK9-induced LDLR degradation. In CHO-K1 cells, overexpression of HLA-C significantly enhanced the degradation of LDLR by exogenous PCSK9. In HepG2 CRISPR HLA-C KO cells, the absence of HLA-C weakened PCSK9's effect on LDLR degradation, while overexpression of HLA-C restored PCSK9's function. These results suggest that HLA-C and HFE may regulate PCSK9's function through different mechanisms, among HFE mainly through inhibiting PCSK9's action on LDLR, while HLA-C through enhancing PCSK9's function to promote LDLR degradation.

Figure 1 Regulatory effects of HLA-C and HFE on PCSK9
The authors used 3D modeling to reveal the importance of HLA-C and PCSK9 interaction and predicted their binding sites at the molecular level. The study found that the R68-G-E70 motif on HLA-C interacts with the M2 domain of PCSK9, while HFE, due to structural similarity, may bind to PCSK9's M2 domain through its R67-V-E69 motif. The interaction between HFE and PCSK9 was further validated using the GRAMM-X Web server for modeling. In experiments, the importance of these sites in the interaction between HFE and PCSK9 was confirmed by mutating or deleting key binding sites on PCSK9 and HFE. Furthermore, the study found that although HFE and HLA-C interact with the same residues of PCSK9, they may encounter PCSK9 through different transport pathways, thus affecting PCSK9's activity on LDLR. These findings provide new perspectives for understanding the interactions between HLA-C, HFE, and PCSK9 and their roles in lipid metabolism.

Figure 2 Direct interaction of PCSK9 with HLA-C compared to HFE
To investigate the internalization pathways of LDL receptor, HLA-C, and HFE and their interactions with PCSK9, the authors used methods to knock down caveolin-1 (siCav1) or clathrin heavy chain (siCHC). The results showed that HLA-C degradation mainly occured through clathrin-coated vesicles, while HFE internalization may depend on the combined action of caveolin and clathrin. The experiments also found that the presence of HFE seemed to inhibit PCSK9's function on LDLR, but further testing using the CircuLex Human PCSK9 Functional Assay Kit showed that the binding affinity between PCSK9 and LDLR remained unchanged regardless of HFE's presence. This suggests that HFE does not compete with LDLR for PCSK9 binding, but rather relies on LDLR's presence to be degraded by PCSK9. Additionally, the lack of LDLR inhibits PCSK9's function on HFE, while HLA-C degradation is not dependent on LDLR. The study also found that the cytoplasmic tail of LDLR is not necessary for PCSK9 to enhance LDLR degradation, while HLA-C plays a key role in sorting the PCSK9-LDLR complex to degradation compartments. These findings help to deepen the understanding of the regulatory mechanisms of LDLR and related proteins in lipid metabolism.

Figure 3 Transport differences between HLA-C and HFE
The authors also used 3D modeling technology to analyze in depth the interactions between PCSK9 and HLA-C and HFE. The results showed that both HLA-C and HFE bind to specific residues E567 and E549 in the CHRD region of PCSK9 through their α1 chains. Although the binding modes of both to PCSK9 are similar, there are subtle differences in the orientation of the α1 chains. At the same time, the N-terminal peptide of PCSK9 has the best interaction with the α1 chain of HLA-C, which was confirmed by Alphafold prediction and modeling. The interaction between the unstructured N-terminal residues of PCSK9 and the peptide-binding pocket of HLA-C shows high confidence, and this binding may be influenced by the structure of PCSK9's N-terminal peptide. Finally, the authors also constructed a complete PCSK9/HLA-C α1 chain/β2-microglobulin ternary model, supporting the view that PCSK9/HLA-C interact through CHRD and N-terminal peptides. This interaction involves residues R68 and E70 on the α1 chain of HLA-C, and residues E567 and E549 on the M2 module of CHRD. In addition, the study also evaluated the similarity of HLA-C antigen pockets to other MHC-I molecules, finding that positively charged residues are generally present in HLA-C antigen pockets, which may support the binding of acidic peptides.

Figure 4 Modeling of interactions between PCSK9 N-terminus and HLA members
Conclusion
This study focuses on the interactions between PCSK9 and LDLR during lysosomal degradation, emphasizing the important roles of PCSK9's M2 domain and HLA-C molecule in this mechanism. It also reveals that HFE and HLA-C exhibit opposite effects in regulating PCSK9 function, and they adopt different endocytosis pathways when interacting with PCSK9. These key findings not only enrich our understanding of protein interactions in cellular transport processes but also provide new directions and perspectives for future research.
Reference
Nabil G. Seidah, et al. “Insights into PCSK9-LDLR Regulation and Trafficking via the Differential Functions of MHC-I Proteins HFE and HLA-C” Cells 5.1 (2024): 13(10): 857


