CRISPR Gene-Editing Breast Cancer Treatment might Change the Face of Modern Medicine

CRISPR Gene-Editing Breast Cancer Treatment might Change the Face of Modern Medicine

The murine mammary carcinoma cell line 4T1 is widely used as a syngeneic tumor model for human breast cancer, a tumor entity with the world-wide highest incidence. 4T1 cells can easily be transplanted into the mammary gland and they are poorly immunogenic, highly tumorigenic, invasive, spontaneously metastasizing to distant organs. Thus, the location of the primary tumor and its metastatic spreading closely resemble the clinical course in patients. Moreover, 4T1 cells are used to investigate triple-negative breast cancer (TNBC) lacking protein expression of estrogen receptor (ER), progesterone receptor (PgR), and epidermal growth factor receptor 2 (ErbB2). This triple-negative phenotype is estimated for more than 17% of breast cancers that are annually diagnosed. Earlier studies reported that mice vaccinated with an EMT6 vaccine exhibited significantly greater protective immunity than mice vaccinated with a 4T1 vaccine. Hybrid vaccination studies revealed that the 4T1 vaccination-induced both local and systemic immune impairments. Thus, 4T1 is among the most popular syngeneic tumor models used for assessing novel therapeutic approaches.
Breast cancer is the second most common cause of death in women in the United States. Most breast cancer deaths are due to metastasis to distal sites of recurrence at the primary site. Breast cancer typically metastasizes to the lung, liver, brain, lymph nodes, and bone. Since one model will not fit all studies, depending on the nature of the study, many murine models are used to mimic the natural process of breast cancer. These models include the tissue or cell inoculation, via ectopic, xenograft, subcutaneous, orthotopic, intra-cardiac.
Researcher choice 4T1 cell line has several characteristics that make it suitable for experiments in the cellular model as well as in animal models for human cancer. It has several advantages over other cancer cell lines because it closely resembles tumor growth and metastasis as seen in human breast cancers. To date, most research on genome editing is designed to better understand diseases by using cultured cells and animal models, while scientists work to gauge the safety and effectiveness of these approaches for human use. Genome editing, and biomarker testing, open the door to patient-tailored therapies that will change outcomes and save lives. This is the future of breast cancer care. CRISPR/Cas9 genome-editing technology has brought invaluable opportunities in the field of breast cancer gene/ cell therapy and could have a crucial role in breast cancer treatment.
Order 4T1 cell line for research→
Application of 4T1 Cell
- 1. Multi-omics studies: genomics, transcriptomics, proteomics, and immunome.
- 2. Cancer recurrence: Identify the cancer recurrence-associated genes, interaction network, downstream studies, and preventive measures of tumor recurrence.
- 3. Gene discovery: Identify new cancer driver genes and uncover cancer-specific vulnerabilities.
- 4. Cancer vaccine: Autologous tumor cell vaccines (ATCVs), Tumor-derived cytokine, ATCV immunogenicity, immunosuppressive cytokines, cancer cell-based vaccines, etc. Study on genetically modified tumor cells for therapeutic cancer vaccine or preventive cancer vaccine.
- 5. Drug discovery: Novel therapeutic vulnerabilities and the development of more effective treatments
- 6. Disease modeling: Allograft and xenograft disease modeling, disease progression, and mechanism studies.
CRISPR-U™ Gene Editing in 4T1 Cell
Genome editing is the process of precisely modifying the nucleotide sequence within a gene for wiping out or initiating explicit and preferred characters in the genome. CRISPR/Cas9 can correct errors in the genome and switch on or off certain genes in cells and organisms fast, cheaply, and relatively easily. 4T1 cell is a model murine carcinoma cell line and is broadly used as syngeneic tumor models for human breast cancer study (drug-resistant gene, chemotherapy-resistant gene, drug discovery, disease mechanisms, initiation, progression, and therapeutics). Gene editing within this cell line possible to generate single or multiple gene knockouts, mutation corrections, or insert reporter transgenes. These genome-edited 4T1 cell lines will allow investigators to study breast cancer hallmarks, disclosure of drug resistance mechanism, cancer therapeutics, cell death research, functional genomics, signaling pathways, drug discovery, drug response, and cell therapy. CRISPR/Cas9 technology positively fuel the advancement of in vivo and in vitro gene editing in breast cancer to tackle the complexity of breast cancer metastasis, drug resistance, chemotherapy resistance and have an immense impact on precision medicine. Ubigene developed CRISPR-U™ for gene manipulation of the 4T1 cell line. Thereby, possible to achieve genome-edited cells with the utilization of the CRISPR/Cas9 system. Ubigene can customize the gene-editing in eukaryotic cells as well as can generate various genes modification in animal models.
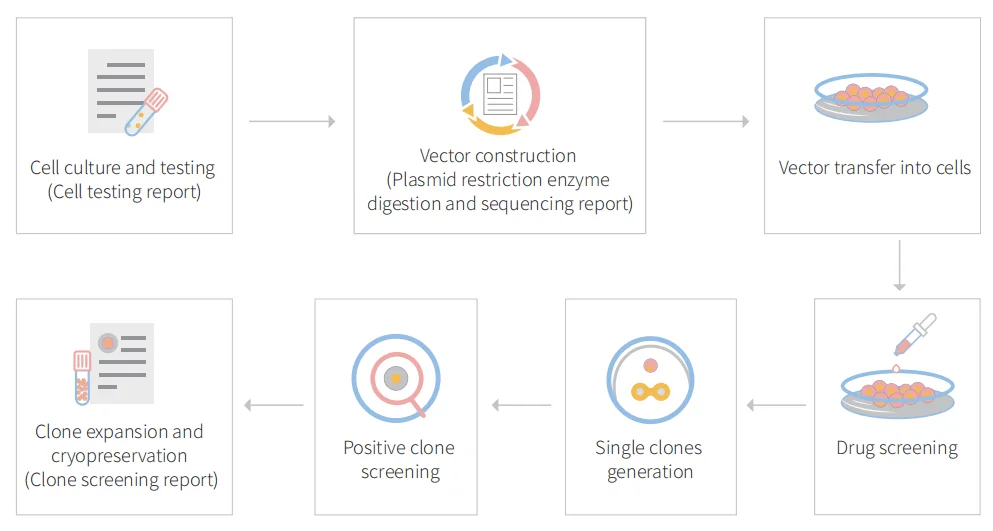
Figure: CRISPR-U™ customized workflow for engineered CT26.WT model cells
Case study of 4T1 cell
CRISPR/Cas9 mediated breast cancer research shows endogenous PAD4 promotes lung metastasis by extracellular chromatin formation
Breast cancer is the most common cancer and the second leading cause of cancer deaths in women in the United States. Along with the aggressive growth of primary tumor mass, the development of distant metastases is a major cause of death from breast cancer. Due to the heterogeneity and lack of well-defined molecular targets, patients with metastatic breast cancer have a poor 5-year survival rate, and finding a method to limit metastasis remains a great challenge. Pathology studies detected the significant expression of PAD4 in many tumor tissues derived from the colon, duodenum, esophagus, fallopian tube, gall bladder, lung, ovary, parotid, pancreas, prostate, rectum, small intestine, stomach, thyroid, and uterus. Peptidyl Arginine Deiminase 4 (PAD4/PADI4) is a posttranslational modification enzyme that converts protein arginine or mono-methylarginine to citrulline.
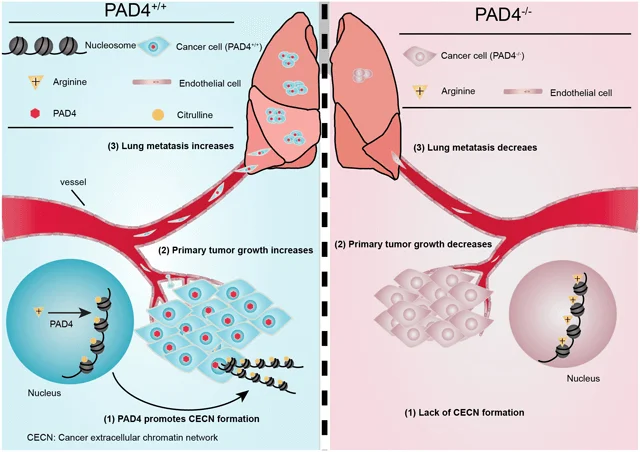
Overview of PAD4 mediated Lung metastasis
To determine whether PAD4 expression was associated with breast cancer in humans, the researcher interrogated the Tissue Cancer Genome Atlas and Oncomine database. After a meta-analysis of gene expression profiling, they found that PAD4 elevated expression was significantly associated with breast carcinoma compared with normal (Fig. 1A). To investigate the functional role of PAD4 in breast cancer, they found highly metastatic mammary carcinoma 4T1 cells express a much higher level of PAD4 than other cancer cell lines (Fig. 1B). They quantify the amount of PADI4 in 4T1 cells and found that PADI4 protein from 5 × 10 4 4T1 cells is roughly comparable to 100 ng purified PAD4 protein, which corresponds to ~16 million molecules of PAD4 protein per cell (Fig. 1C).
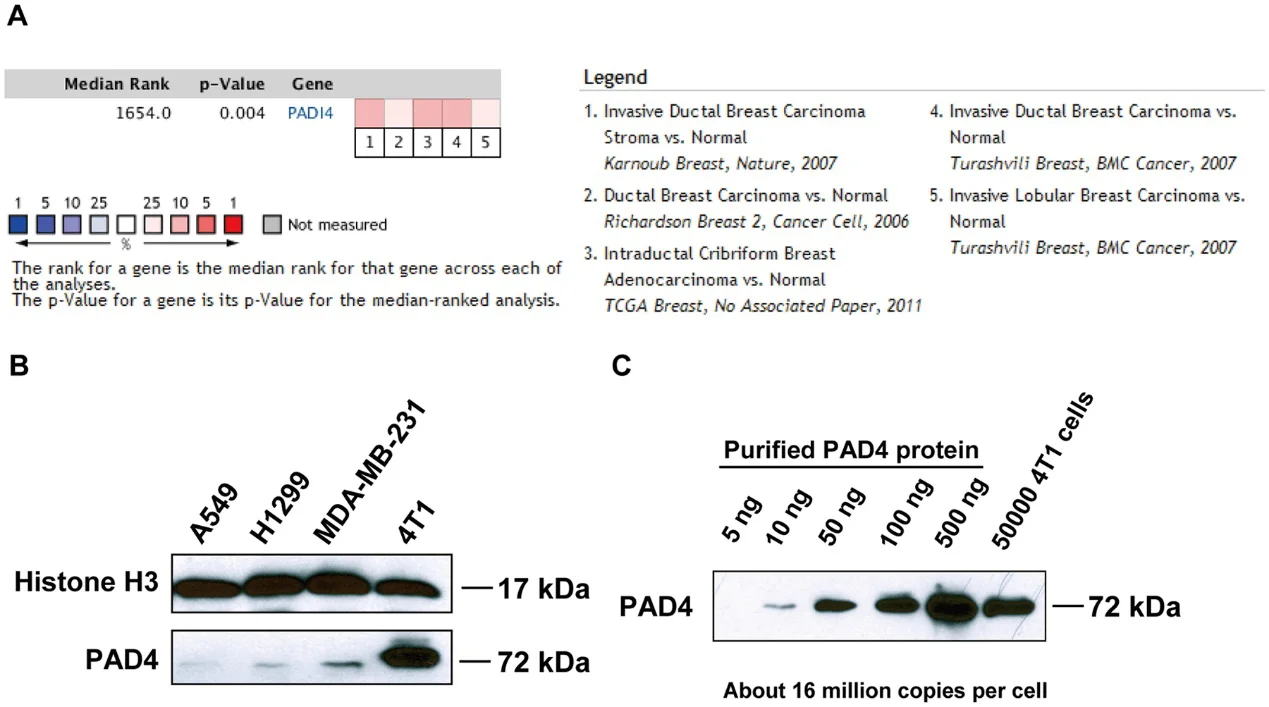
Fig 1: PAD4 expression in human and mouse mammary tumors.
PAD4 is a calcium-dependent enzyme. Therefore, they performed Western blot experiments after calcium ionophore treatment and found that histone H3 citrullination detected with the H3Cit antibody (made against a histone H3 N-terminal peptide containing Cit2, Cit8, and Cit17) increased significantly after calcium ionophore treatment (Fig. 2A). To test how histone citrullination activity affects chromatin structure they analyzed cell morphology with immunostaining, H3Cit staining was greatly increased at areas of highly decondensed chromatin (Fig. 2B). They tested the citrullination level on two positions with α-H3Cit26 (Fig. 2C and 2D) and α-H4Cit3 antibodies (Fig. 2E and 2F) respectively. To further confirm the presence of extracellular chromatin structure, intact 4T1 cells with or without calcium ionophore treatment were subjected to digestion with MNase, a nuclease that cuts between nucleosomes in chromatin (Fig. 2G). Accordingly, histone H3 protein can only be detected from the supernatant of 4T1 cells after calcium ionophore stimulation (Fig. 2H). Together, they found a modest increase in extracellular DNA and H3Cit staining after exposure of 4T1 cells to HMGB1, indicating the ability of HMGB1 to induce CECN in 4T1 cells (Fig. 2I and 2J).
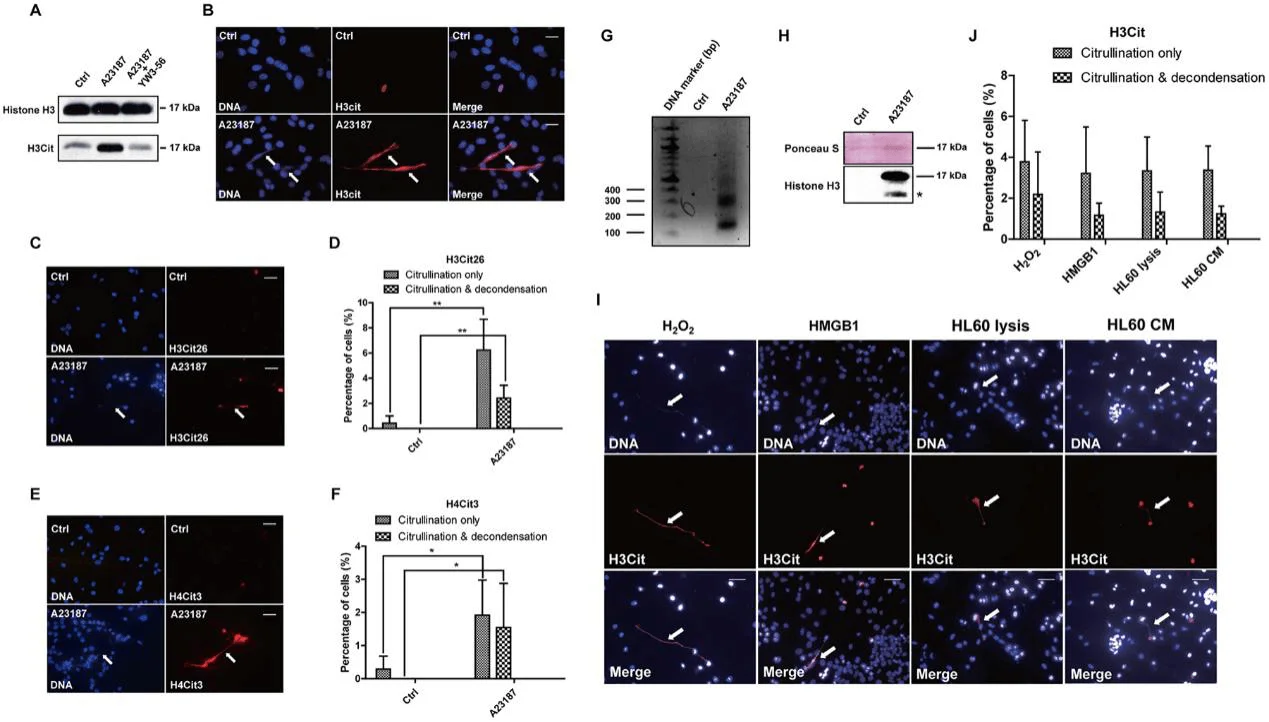
Fig 2: Activation of PADI4 leads to hyper-citrullination and CECN formation in 4T1 cells in-vitro.
They found that inhibition of PAD decreased the incidence of both hyper-citrullination and release of CECN (Fig. 3A). They knocked out Padi4 in tumor cells using the CRISPR/Cas9 method and selected two single-cell colonies named Padi4 CRISPR-Ex1–1-C11 and Padi4 CRISPR-Ex2–6-E2 with Padi4 deletion in exon 1 and 2, respectively. The deletion of Padi4 was confirmed by Sanger Sequencing, and Western blot (Fig. 3B, C). Citrullination and the formation of CECN were not detected after calcium ionophore stimulation (Fig. 3D). Thus, PAD4 activity is required for histone hyper-citrullination and CECN formation.
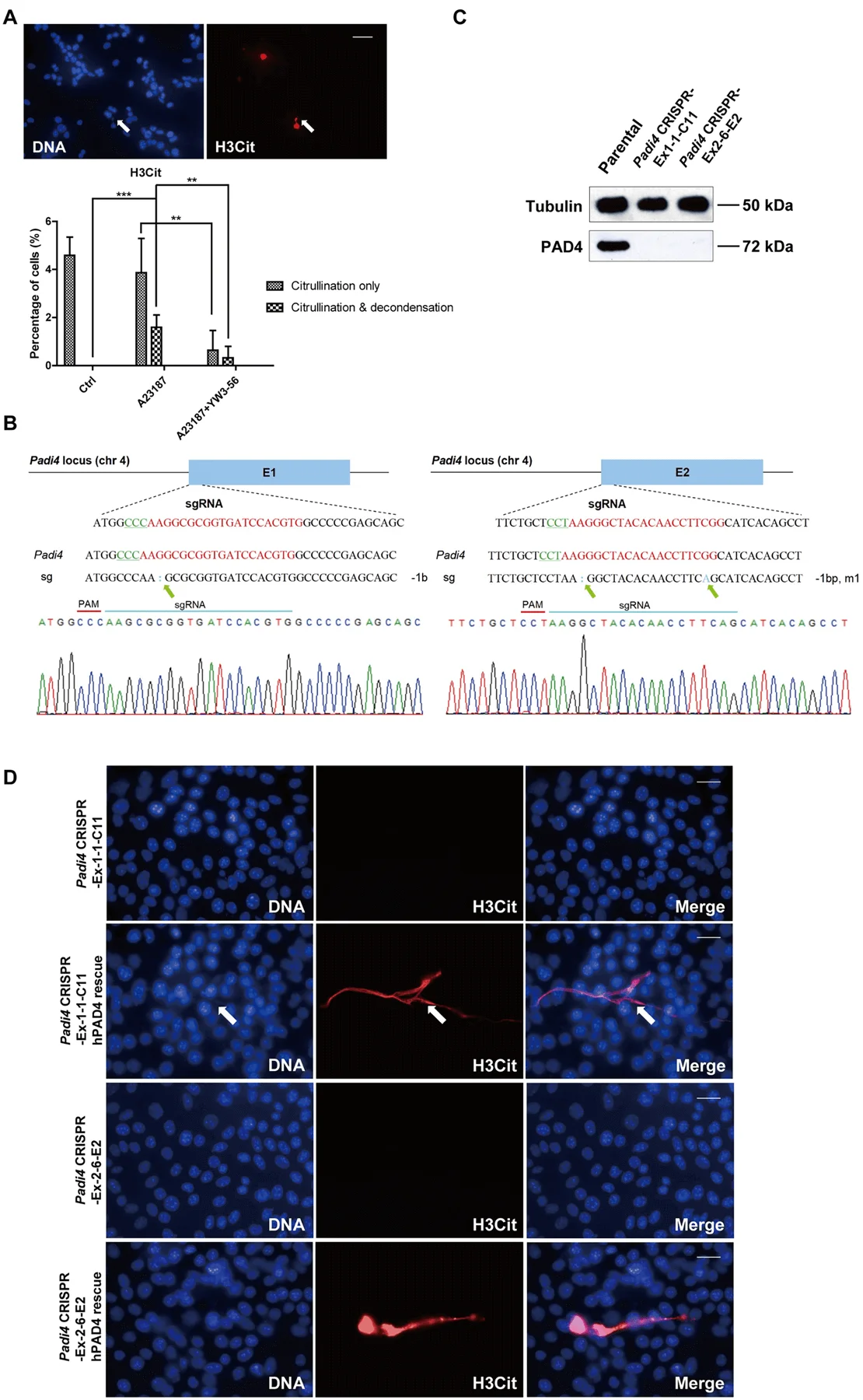
Fig 3: Formation of CECN is dependent on PAD4.
To investigate whether 4T1 cells can release CECN in vivo, they compared citrullination levels from orthotopically transplanted and size-matched 4T1 wild type and Padi4 knockout tumors. PAD4 was only detected in the parental cell line and parental tumors (Fig. 4A). To test whether hyper-citrullination leads to decondensed chromatin are shown in Fig. 4B. Immunostained were performed to detect potential CECN structure (Fig. 4C). H&E staining and immunostaining experiments with tumors allografted on BALB/c Padi4 knockout mice are shown in Fig. 4D to 4F. The H3Cit staining in the primary tumor is area-based and has different staining patterns, with some areas positive while others negative of H3Cit staining (Fig. 4G). This is likely due to the heterogeneity of tumor microenvironment with different levels of ROS or numbers of dying cells.
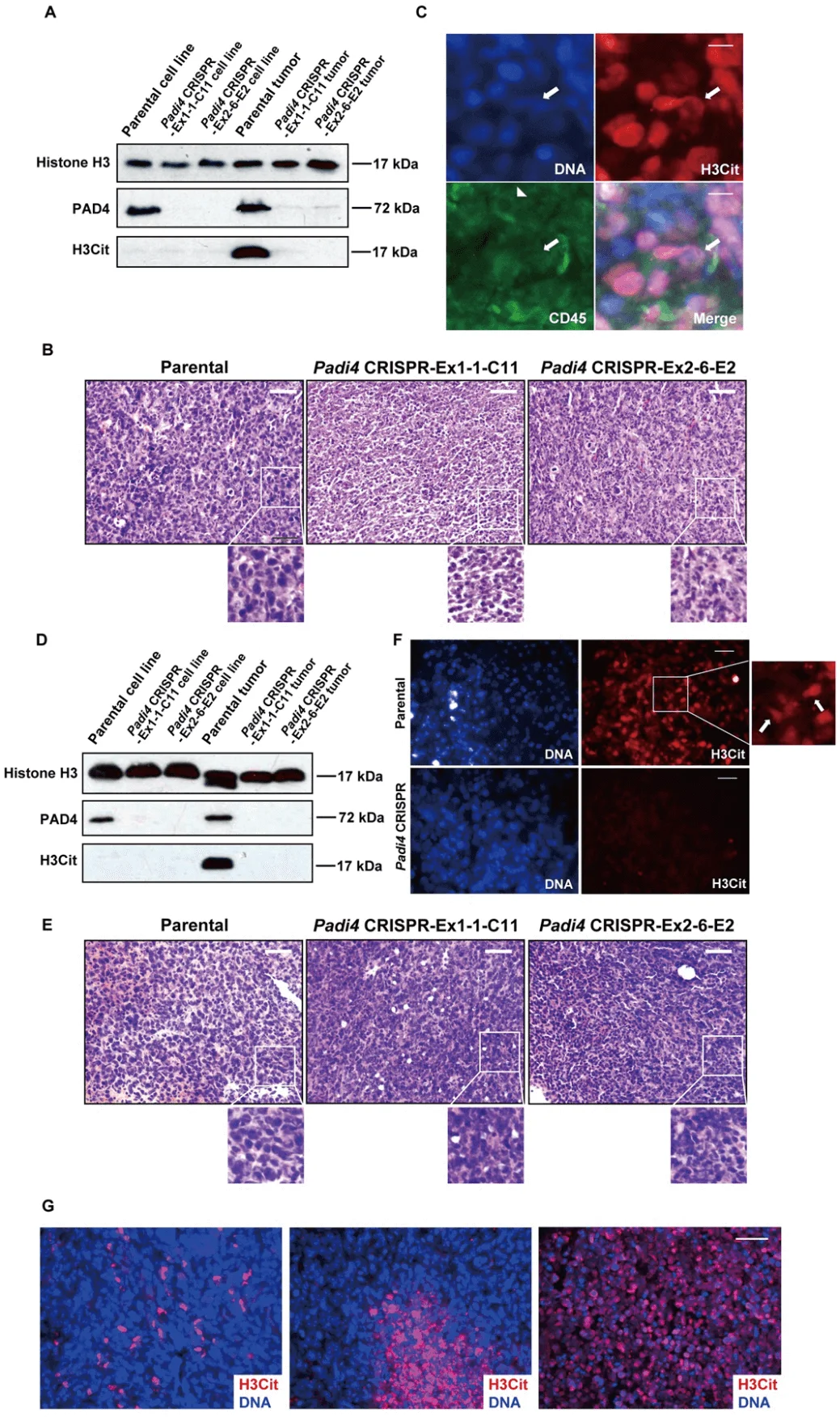
Fig 4: CECN is present in allograft tumors.
To assess the cell-autonomous role of PAD4 on tumor growth and metastasis, wild type female BALB/c mice were inoculated in the mammary gland with 4T1 wild type and Padi4 knockout cells. Although all 4T1 cell types showed similar degrees of in vitro growth rates (Fig. 5A), the allograft Padi4 knockout tumors grew significantly slower compared with wild type 4T1 tumors in wild type BALB/c mice (Fig. 5B). The reduced growth rate of Padi4 knockout tumors was also observed in Padi4 knockout BALB/c mice (Fig. 5C). Similarly, intratumoral PAD4 inhibitor treatment also reduced the primary tumor growth rate (Fig. 5D).
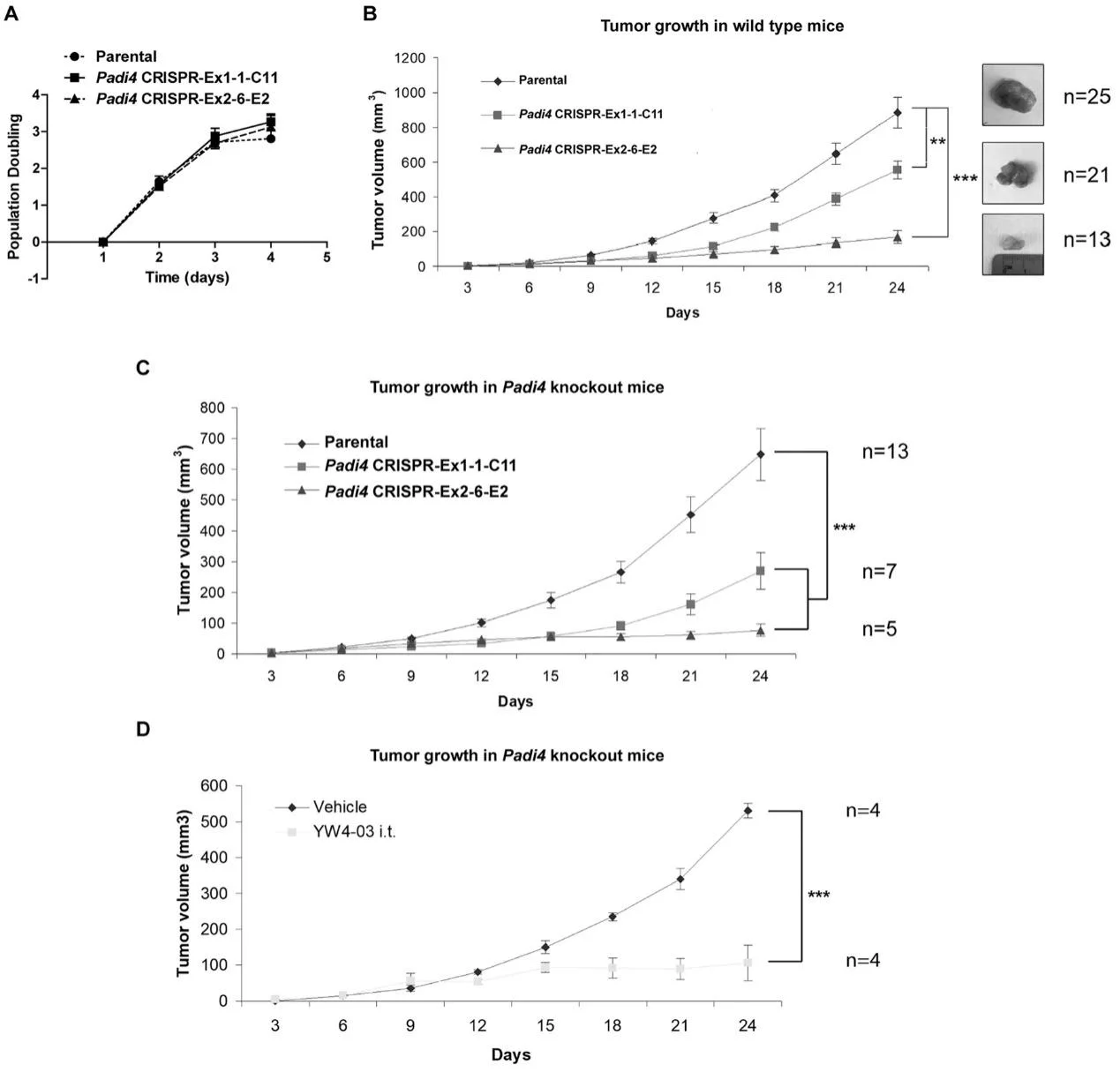
Fig 5: Growth rates of primary mammary tumors formed by 4T1 cells with different Padi4 genotypes or combined with YW4–03 treatment.
4T1 cells are highly metastatic compared with a sibling cell line 67NR which was derived from the same mouse tumor but cannot metastasize. As shown in Fig. 6A, the PAD4 level is significantly higher in 4T1 cells compared with 67NR cells. After allograft, mice bearing wild type tumors formed a significantly higher number of metastatic foci on the lung surface than the two Padi4 knockout cell lines (Fig. 6B and 6C). Lung metastasis was still significantly reduced in mice bearing tumors derived from Padi4 CRISPR cancer cells even after prolonged growth (Fig. 6D–6F). To test the effects of PADI4 inhibition on a population level, lung metastasis was also compared in the YW4–03 intratumoral treatment model using Padi4 knockout mice (Fig. 6G and 6H).

Fig 6: Lack of PAD4 in tumors leads to reduced lung metastasis.
To further test the idea that CECNs are involved in metastasis, they used DNase I to disrupt extracellular DNA including CECN structures in the 4T1 wild-type tumors growing in the Padi4 knockout mice, lung metastasis was significantly decreased (Fig. 7A and 7B). Also, tested whether PAD4 expressed in cancer cells influences intravasation by analyzing the Circulating Tumor Cells (CTCs) in blood with colony formation assays. Neither loss of Padi4 nor DNase I treatment influenced the CTC numbers from size-matched tumor-bearing mice, suggesting cancer cell endogenous PAD4 does not promote metastasis through the intravasation process (Fig. 7C–7F). To further test whether PADI4 is functioning at steps after intravasation, they performed experimental metastasis assays by intravenously injecting the same number of tumor cells into mice. The loss of Padi4 in 4T1 cells didn’t significantly reduce the number of cancer cells accumulated compared with the Padi4 wild type cells (Fig. 7G and 7H).
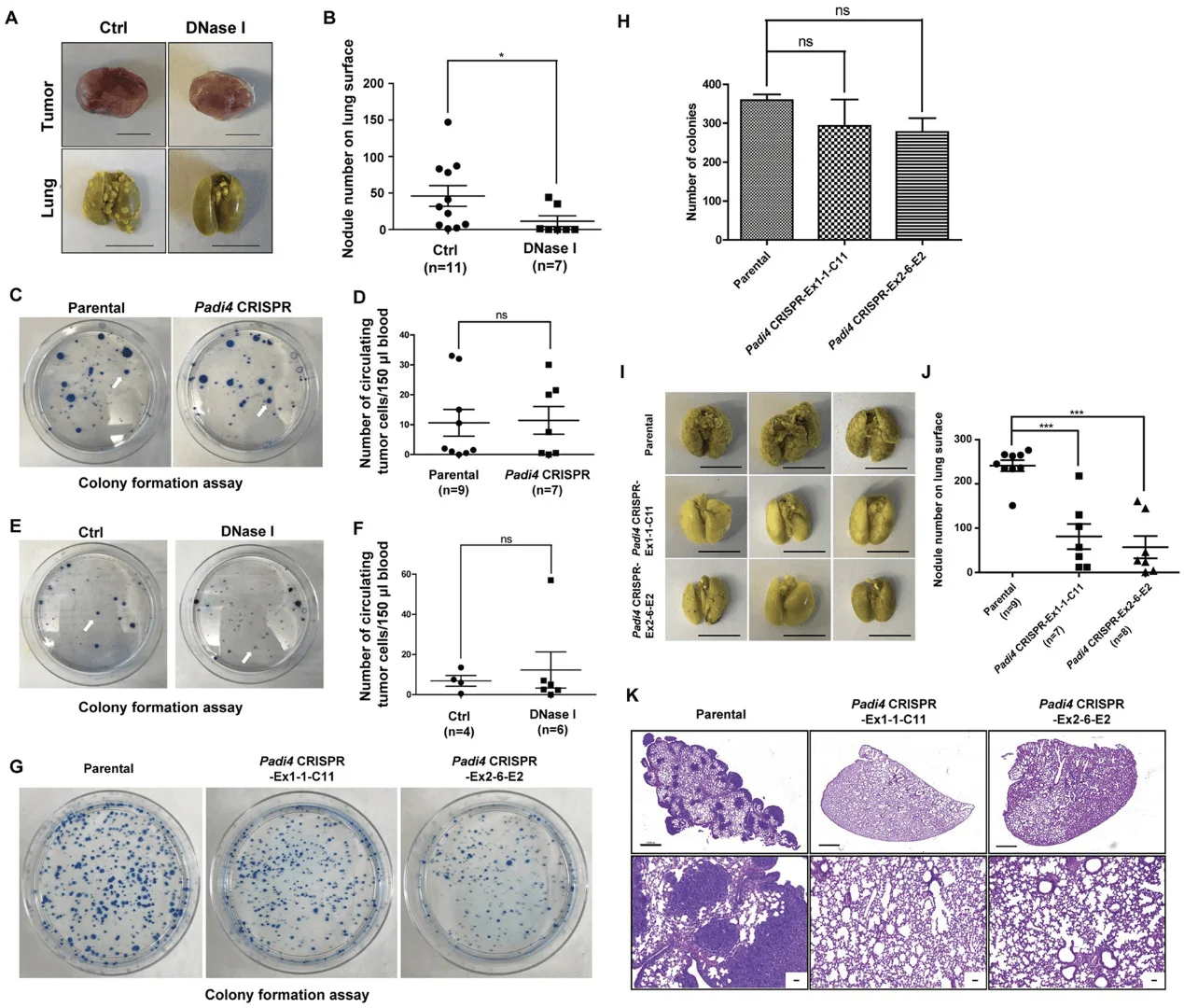
Fig 7: Cancer cell endogenous PAD4 promotes metastasis through steps after intravasation.
In summary, the researcher establishes the occurrence of PADI4-mediated CECN in 4T1 cells in vitro and in vivo. The tumor microenvironment harbors the signal to activate PADI4 and deletion of Padi4 or removal of extracellular DNA including CECN with DNase I in 4T1 cells inhibits metastatic lung colonization. PAD4’s vital role in promoting tumor growth and metastasis renders itself a tangible therapeutic.
Ubigene developed CRISPR-U™ which optimizes eukaryotic cells and animal gene-editing vectors and processes. The efficiency and accuracy are 10x higher than traditional methods. Contact us immediately to know about your research related services!

Reference
Endogenous PAD4 in Breast Cancer Cells Mediates Cancer Extracellular Chromatin Network Formation and Promotes Lung Metastasis Mol Cancer Res . 2020; 18(5): 735–747.
You may also interested in:
Popular Breast Cancer Cell Models Unveiled!
IF=16.6| 4T1 Luciferase Cell Aids in the Study of Disseminated Breast Cancer Metastasis


