

Location:Home > Application > H1299 cell, an ideal route for cancer mutation CRISPR therapy research
H1299 cell, an ideal route for cancer mutation CRISPR therapy research

Lung cancer is the leading cause of cancer-related deaths worldwide, with a 5-year survival of approximately 6%. Around 80% of these are of non-small-cell (NSCLC) histological type for which surgical resection or radical chemoradiotherapy offers the best prospect of cure. Many lung cancer patients are resistant to current treatments, including chemotherapy and radiotherapy. The vast majority of cases however are diagnosed at an advanced stage, and therapy options are limited. Despite progress in research on lung cancer, targeted therapeutics, and clinical management, many knowledge gaps remain to be closed. Due to lung cancer's genetic and phenotypic diversity, individualization of therapy is becoming a reality; thus, this tumor entity is likely to further trigger future precision medicine development.
Nonsmall cell lung cancer (NSCLC) model cell line, H1299 is widely used in a variety of basic cell biology and biomedical studies involving lung cancer proliferation, corresponding inhibitors studies, tumor formation, and metastases. The researcher uses H1299 cell line as a disease model to understand the basic biology of the disease, to understand how mutations affect drug response or resistance, to understanding the mechanisms underlying differences in drug responsiveness, target identification, and validation as well as even to stratify patients for more efficient and effective clinical trials. Gene editing technologies are rapidly advancing as a realistic therapeutic option. The ability to strategically edit a patient’s genome can constitute a treatment revolution. Genome editing technologies have huge potential in lung cancer treatment including targeting oncogenes and tumor-suppressor genes, genes related to chemotherapy drug resistance, and genes related to therapies using targeted drugs and inhibitors to promote further preclinical research and the clinical treatment of lung cancer.
H1299 cell line (NCI-H1299 or CRL-5803) was established from the lung cells of a 43-year-old Caucasian male patient with non-small cell lung cancer and is widely used in biomedical research. An immortalized cell line, H1299 can divide indefinitely and the unique feature of this cell line is the lack of expression of the P53 protein, which is accounts for their proliferative propensity. This cell line (H1299) has been reported to secrete the peptide hormone neuromedin B(NMB), but not gastrin-releasing peptide (GRP) and are useful for studying lung cancer in humans. An earlier study reported that the H1299 cell line used to study genes related to sensitization and drug resistance to lung cancer chemotherapeutics such as paclitaxel, providing a theoretical basis for improving the therapeutic efficacy of chemotherapy in lung cancer.
Below are some applications of H1299:
1.Targeted therapy: Anti-angiogenetic drugs, Drugs targeting cell signaling, biologic mechanism, toxicity profile, Other biologic agents (vandetanib, sorafenib, etc.).
2. Drug developments: PI3K inhibitors, MEK inhibitors, EGFR inhibitors, etc.
3. Molecular genetics: Exploitation of the genomic aberrations, somatic genetic alterations, oncogenic activation of particular tyrosine kinases.
4. Epigenetics: Mutations in epigenetic regulators, epigenetic therapy, etc.
CRISPR-U™ gene editing in H1299 cell line
Genome editing and the creation of cellular models can advance research programs in the area of functional genomics, signaling pathways, metabolism, cell death, drug discovery, drug response, and cancer research, etc. The CRISPR system is a precise genome editing technique, which can create gene knockout or knock-in genome manipulations through the substitution of a target genetic sequence with a desired donor sequence. H1299 is a model human lung adenocarcinoma cell line, used for lung cancer study mainly drug discovery, disease mechanisms, initiation, progression, and therapeutics. CRISPR-system mediated edited H1299 cell lines allow investigators to study cancer hallmarks, disclosure of drug resistance mechanism, cancer therapeutics, cell death research, functional genomics, signaling pathways, drug discovery, drug response, and cell therapy. CRISPR/Cas9 technology positively fuel the advancement of in vivo and in vitro gene editing in lung cancer and have an immense impact on molecular medicine. Besides, knockout of the RSF1 gene in combination with paclitaxel resulted in the cell-cycle arrest in G1, increased apoptosis, and reduced cell migration and proliferation. Also, Knocking out NESTIN in H1299 cells can promote apoptosis, inhibit proliferation and colony formation, and suppress cell invasion by inhibiting epithelial-to-mesenchymal transition (EMT). Ubigene developed CRISPR-U™ for gene manipulation of the H1299 cell line. Thereby, possible to achieve genome-edited cells with the utilization of the CRISPR/Cas9 system. Ubigene can customize the gene-editing in eukaryotic cells as well as can generate various genes modification in animal models.
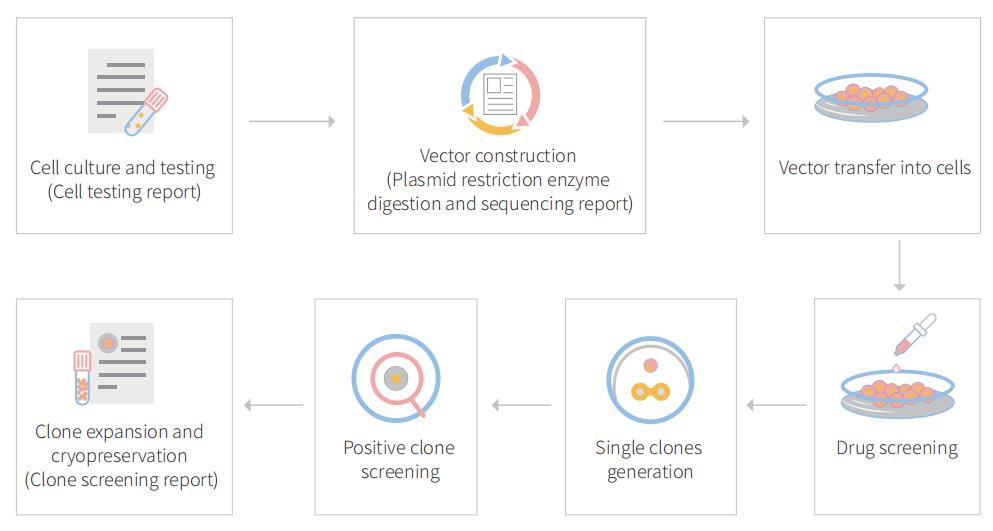
Figure: CRISPR-U™ customized workflow for engineered H1299 model cells
BCAR1 knockout cell line reveals that BCAR1 promotes proliferation and cell growth in lung adenocarcinoma via upregulation of POLR2A
Breast cancer antiestrogen resistance protein 1 (BCAR1; Crk‐associated substrate, CAS; p130cas) directly interact with the multiple protein motifs of various phosphatases and kinases, and to mediate Src through FAK bridge indirect association. BCAR1 has been reported to enhance tumor proliferation, invasion, and metastasis in several cancers (prostate cancer, endometrial adenocarcinoma, oral squamous cell carcinoma, breast cancer, and lung cancer). it also predicts poorer prognosis in lung adenocarcinoma cases.
In this study, the researcher explores the role of BCAR1 in proliferation and cell growth in lung adenocarcinoma and the networks of proteins that interact with BCAR1 to trigger the proliferation of lung cancer cells. They used to lung adenocarcinoma cell line (NCl-H1975 and NCl-H1299) and established BCAR1 knockout cell line by CRISPR/Cas9 system as well as performed KO cell line for cell proliferation, colony formation, apoptosis, and cell cycle assay (Fig.1) They found that BCAR1 expression in the KO group was significantly lower than that of control as well as proliferation was significantly inhibited (Fig.1 b,c). Colony‐formation efficiency of H1975 cells was significantly decreased but no significant difference between BCAR‐KO and NC in H1299 cells (Fig.1d). Apoptosis or cell cycle progression analysis revealed that no difference in apoptosis and cell cycle of H1975 and H1299 cells following BCAR‐KO (Fig.1 e,f).
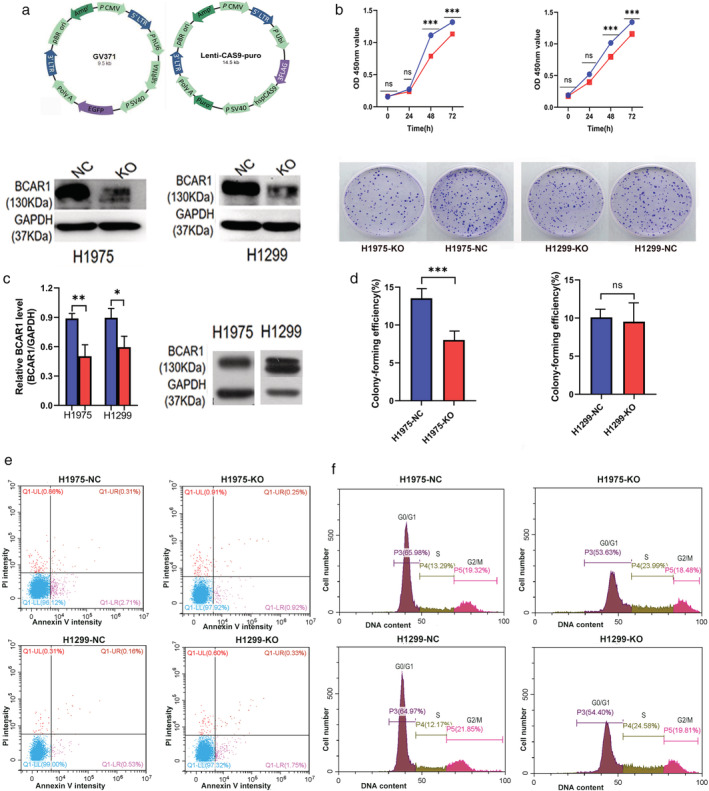
Figure 1: Cell proliferation, colony formation, apoptosis, and cell cycles of H1975 and H1299 cell after BCAR1 knockout
They overexpressed BCAR1 in 293T cell lines and performed immune precipitation-mass spectrometric (IP-MS) as well as different downstream analysis. The IP-MS and bioinformatic analysis predict potential interactions with BCAR1 (Fig. 2). Bioinformatic analysis revealed potential BCAR1 interact proteins and interaction partners catalytic activity and transferase activity (Fig.2a,b). The Cancer Genome Atlas (TCGA) database verification confirms BCAR1 overexpression correlates with cancer (Fig.2c). They analyze cancer-related interacting partner PPI analysis and found that high expression of POLR2A, MAPK3, MOV10, and XAB2 predicted poor prognosis in lung adenocarcinoma (Fig.2d). They consider POLR2A for further verification due to the link with more genes and the possibility of involvement in the signaling cascade.

Figure-2: Bioinformatic analysis of BCAR1 overexpressed in 293cell line
POLR2A and BCAR1 were significantly increased in lung adenocarcinoma tissues compared to adjacent normal tissues (Fig.3a). IHC‐stained TMA is shown in Fig 3b, demonstrate that BCAR1 was expressed in the nucleus, in the cytoplasm, or both locations (Fig.3c). However, POLR2A was highly expressed in the nucleus. POLR2A expression was significantly positively correlated to BCAR1 expression (R = 0.476, P < 0.001). Neither BCAR1 nor POLR2A expression was correlated with tumor size. High expression of either BCAR1 or POLR2A predicted poor prognosis in 54 lung cancer cases in the early-stage (Fig. 3d). They further verified POLR2A in H1975, and H1299 cell KO clone and western blot analysis demonstrated that POLR2A was significantly decreased in BCAR1‐KO compared to NC cells (Fig.3e). BCAR1 regulates POLR2A in H1975 and H1299 cells, despite the negative results of the CO‐IP (Fig.3f).
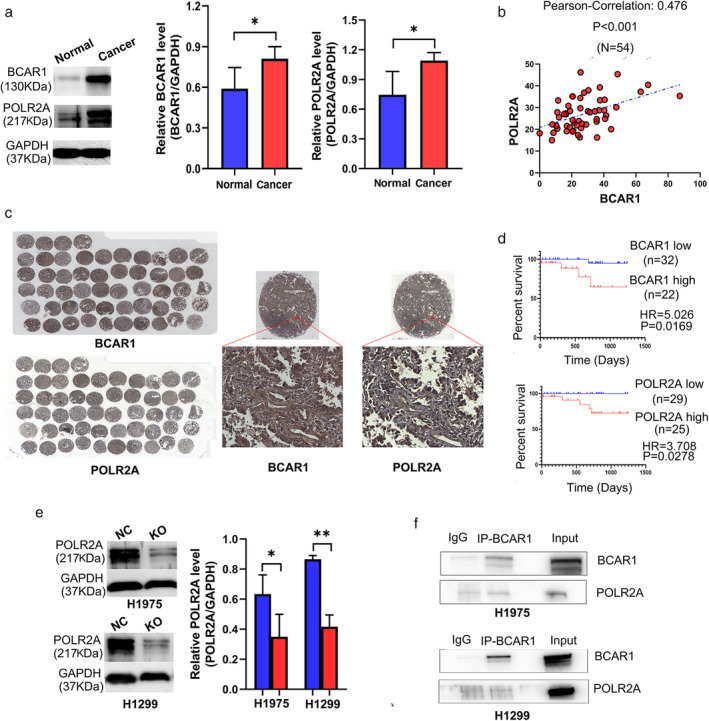
Figure-3: The relationship between BCAR1 and POLR2A as well as their prognostic significance in lung adenocarcinoma.
In summary, the researcher did not find a direct interaction between BCAR1 and POLR2A in H1975 and H1299 cells. The underlying mechanism of how BCAR1 and POLR2A are connected remains unclear. Moreover, synergism analysis is needed to demonstrate the role of interaction between BCAR1 and its partners concerning proliferation and cell growth in lung cancer. Future robust studies are required to resolve the interesting abovementioned points.
Ubigene developed CRISPR-U™ which optimizes eukaryotic cells and animal gene-editing vectors and processes. The efficiency and accuracy are 10x higher than traditional methods. Contact us immediately to know about your research related services!

Reference:
BCAR1 promotes proliferation and cell growth in lung adenocarcinoma via upregulation of POLR2A.Thorac Cancer. 2020; 11(11): 3326–3336.


H1299 cell, an ideal route for cancer mutation CRISPR therapy research

Lung cancer is the leading cause of cancer-related deaths worldwide, with a 5-year survival of approximately 6%. Around 80% of these are of non-small-cell (NSCLC) histological type for which surgical resection or radical chemoradiotherapy offers the best prospect of cure. Many lung cancer patients are resistant to current treatments, including chemotherapy and radiotherapy. The vast majority of cases however are diagnosed at an advanced stage, and therapy options are limited. Despite progress in research on lung cancer, targeted therapeutics, and clinical management, many knowledge gaps remain to be closed. Due to lung cancer's genetic and phenotypic diversity, individualization of therapy is becoming a reality; thus, this tumor entity is likely to further trigger future precision medicine development.
Nonsmall cell lung cancer (NSCLC) model cell line, H1299 is widely used in a variety of basic cell biology and biomedical studies involving lung cancer proliferation, corresponding inhibitors studies, tumor formation, and metastases. The researcher uses H1299 cell line as a disease model to understand the basic biology of the disease, to understand how mutations affect drug response or resistance, to understanding the mechanisms underlying differences in drug responsiveness, target identification, and validation as well as even to stratify patients for more efficient and effective clinical trials. Gene editing technologies are rapidly advancing as a realistic therapeutic option. The ability to strategically edit a patient’s genome can constitute a treatment revolution. Genome editing technologies have huge potential in lung cancer treatment including targeting oncogenes and tumor-suppressor genes, genes related to chemotherapy drug resistance, and genes related to therapies using targeted drugs and inhibitors to promote further preclinical research and the clinical treatment of lung cancer.
H1299 cell line (NCI-H1299 or CRL-5803) was established from the lung cells of a 43-year-old Caucasian male patient with non-small cell lung cancer and is widely used in biomedical research. An immortalized cell line, H1299 can divide indefinitely and the unique feature of this cell line is the lack of expression of the P53 protein, which is accounts for their proliferative propensity. This cell line (H1299) has been reported to secrete the peptide hormone neuromedin B(NMB), but not gastrin-releasing peptide (GRP) and are useful for studying lung cancer in humans. An earlier study reported that the H1299 cell line used to study genes related to sensitization and drug resistance to lung cancer chemotherapeutics such as paclitaxel, providing a theoretical basis for improving the therapeutic efficacy of chemotherapy in lung cancer.
Below are some applications of H1299:
1.Targeted therapy: Anti-angiogenetic drugs, Drugs targeting cell signaling, biologic mechanism, toxicity profile, Other biologic agents (vandetanib, sorafenib, etc.).
2. Drug developments: PI3K inhibitors, MEK inhibitors, EGFR inhibitors, etc.
3. Molecular genetics: Exploitation of the genomic aberrations, somatic genetic alterations, oncogenic activation of particular tyrosine kinases.
4. Epigenetics: Mutations in epigenetic regulators, epigenetic therapy, etc.
CRISPR-U™ gene editing in H1299 cell line
Genome editing and the creation of cellular models can advance research programs in the area of functional genomics, signaling pathways, metabolism, cell death, drug discovery, drug response, and cancer research, etc. The CRISPR system is a precise genome editing technique, which can create gene knockout or knock-in genome manipulations through the substitution of a target genetic sequence with a desired donor sequence. H1299 is a model human lung adenocarcinoma cell line, used for lung cancer study mainly drug discovery, disease mechanisms, initiation, progression, and therapeutics. CRISPR-system mediated edited H1299 cell lines allow investigators to study cancer hallmarks, disclosure of drug resistance mechanism, cancer therapeutics, cell death research, functional genomics, signaling pathways, drug discovery, drug response, and cell therapy. CRISPR/Cas9 technology positively fuel the advancement of in vivo and in vitro gene editing in lung cancer and have an immense impact on molecular medicine. Besides, knockout of the RSF1 gene in combination with paclitaxel resulted in the cell-cycle arrest in G1, increased apoptosis, and reduced cell migration and proliferation. Also, Knocking out NESTIN in H1299 cells can promote apoptosis, inhibit proliferation and colony formation, and suppress cell invasion by inhibiting epithelial-to-mesenchymal transition (EMT). Ubigene developed CRISPR-U™ for gene manipulation of the H1299 cell line. Thereby, possible to achieve genome-edited cells with the utilization of the CRISPR/Cas9 system. Ubigene can customize the gene-editing in eukaryotic cells as well as can generate various genes modification in animal models.

Figure: CRISPR-U™ customized workflow for engineered H1299 model cells
BCAR1 knockout cell line reveals that BCAR1 promotes proliferation and cell growth in lung adenocarcinoma via upregulation of POLR2A
Breast cancer antiestrogen resistance protein 1 (BCAR1; Crk‐associated substrate, CAS; p130cas) directly interact with the multiple protein motifs of various phosphatases and kinases, and to mediate Src through FAK bridge indirect association. BCAR1 has been reported to enhance tumor proliferation, invasion, and metastasis in several cancers (prostate cancer, endometrial adenocarcinoma, oral squamous cell carcinoma, breast cancer, and lung cancer). it also predicts poorer prognosis in lung adenocarcinoma cases.
In this study, the researcher explores the role of BCAR1 in proliferation and cell growth in lung adenocarcinoma and the networks of proteins that interact with BCAR1 to trigger the proliferation of lung cancer cells. They used to lung adenocarcinoma cell line (NCl-H1975 and NCl-H1299) and established BCAR1 knockout cell line by CRISPR/Cas9 system as well as performed KO cell line for cell proliferation, colony formation, apoptosis, and cell cycle assay (Fig.1) They found that BCAR1 expression in the KO group was significantly lower than that of control as well as proliferation was significantly inhibited (Fig.1 b,c). Colony‐formation efficiency of H1975 cells was significantly decreased but no significant difference between BCAR‐KO and NC in H1299 cells (Fig.1d). Apoptosis or cell cycle progression analysis revealed that no difference in apoptosis and cell cycle of H1975 and H1299 cells following BCAR‐KO (Fig.1 e,f).

Figure 1: Cell proliferation, colony formation, apoptosis, and cell cycles of H1975 and H1299 cell after BCAR1 knockout
They overexpressed BCAR1 in 293T cell lines and performed immune precipitation-mass spectrometric (IP-MS) as well as different downstream analysis. The IP-MS and bioinformatic analysis predict potential interactions with BCAR1 (Fig. 2). Bioinformatic analysis revealed potential BCAR1 interact proteins and interaction partners catalytic activity and transferase activity (Fig.2a,b). The Cancer Genome Atlas (TCGA) database verification confirms BCAR1 overexpression correlates with cancer (Fig.2c). They analyze cancer-related interacting partner PPI analysis and found that high expression of POLR2A, MAPK3, MOV10, and XAB2 predicted poor prognosis in lung adenocarcinoma (Fig.2d). They consider POLR2A for further verification due to the link with more genes and the possibility of involvement in the signaling cascade.

Figure-2: Bioinformatic analysis of BCAR1 overexpressed in 293cell line
POLR2A and BCAR1 were significantly increased in lung adenocarcinoma tissues compared to adjacent normal tissues (Fig.3a). IHC‐stained TMA is shown in Fig 3b, demonstrate that BCAR1 was expressed in the nucleus, in the cytoplasm, or both locations (Fig.3c). However, POLR2A was highly expressed in the nucleus. POLR2A expression was significantly positively correlated to BCAR1 expression (R = 0.476, P < 0.001). Neither BCAR1 nor POLR2A expression was correlated with tumor size. High expression of either BCAR1 or POLR2A predicted poor prognosis in 54 lung cancer cases in the early-stage (Fig. 3d). They further verified POLR2A in H1975, and H1299 cell KO clone and western blot analysis demonstrated that POLR2A was significantly decreased in BCAR1‐KO compared to NC cells (Fig.3e). BCAR1 regulates POLR2A in H1975 and H1299 cells, despite the negative results of the CO‐IP (Fig.3f).

Figure-3: The relationship between BCAR1 and POLR2A as well as their prognostic significance in lung adenocarcinoma.
In summary, the researcher did not find a direct interaction between BCAR1 and POLR2A in H1975 and H1299 cells. The underlying mechanism of how BCAR1 and POLR2A are connected remains unclear. Moreover, synergism analysis is needed to demonstrate the role of interaction between BCAR1 and its partners concerning proliferation and cell growth in lung cancer. Future robust studies are required to resolve the interesting abovementioned points.
Ubigene developed CRISPR-U™ which optimizes eukaryotic cells and animal gene-editing vectors and processes. The efficiency and accuracy are 10x higher than traditional methods. Contact us immediately to know about your research related services!

Reference:
BCAR1 promotes proliferation and cell growth in lung adenocarcinoma via upregulation of POLR2A.Thorac Cancer. 2020; 11(11): 3326–3336.




