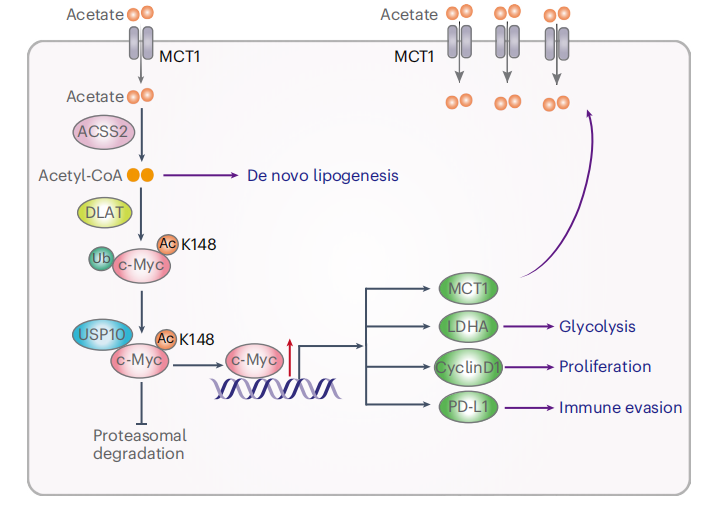
Location: Home > Gene Editing Services > Stable Cell Lines > knockin cell line


| Cell type | Various types of cells including tumor cell lines, regular cell lines, IPS/ES cell lines |
| Service type | RNP Method, Prime Editor, Base Editing, Antibiotics-based Konck in |
| Deliverables | Single-cell clone |
| Turnaround/Price | 8 weeks,as low as $6480 |
300+Successful
Gene-editing Cell Line Types
 Respiratory System
Respiratory System Circulatory System
Circulatory System Endocrine System
Endocrine System Brain and Nervous System
Brain and Nervous System Blood and lymphatic System
Blood and lymphatic System Urinary System
Urinary System Digestive System
Digestive System Skeleton, Articulus, Soft Tissue, Derma System
Skeleton, Articulus, Soft Tissue, Derma System Ocular, Otolaryngologic and Oral System
Ocular, Otolaryngologic and Oral System Reproductive System
Reproductive System Stem Cell
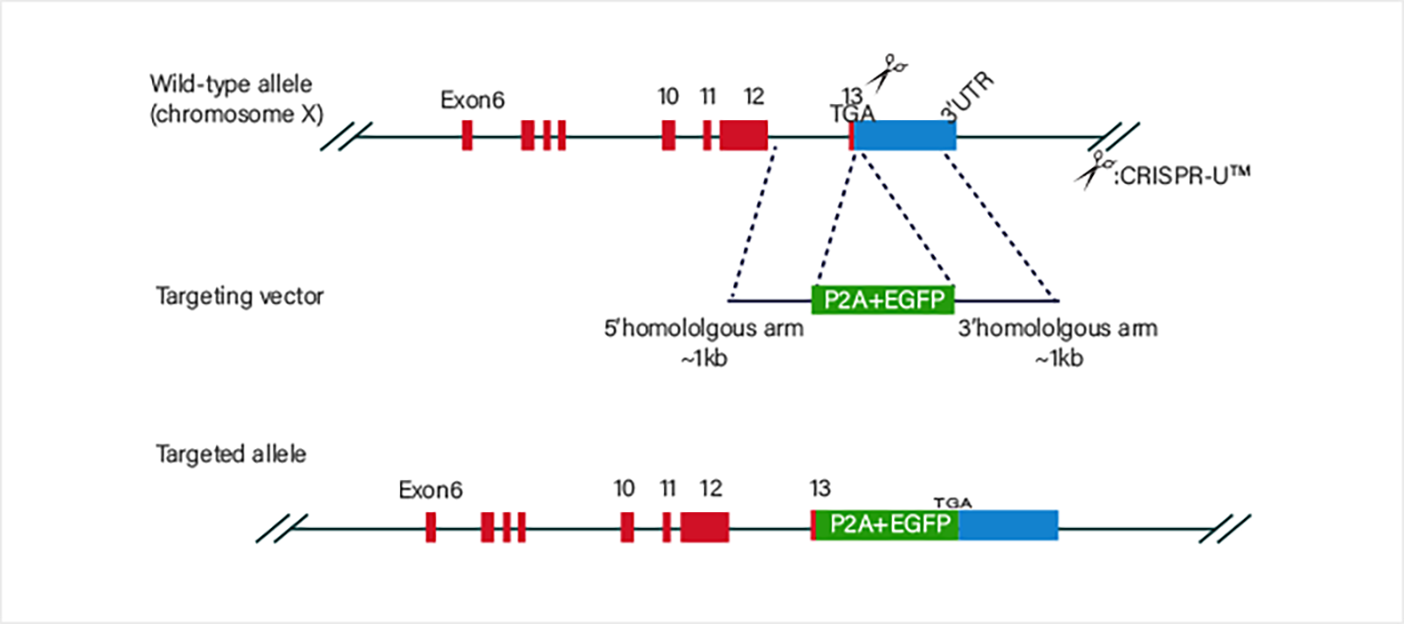
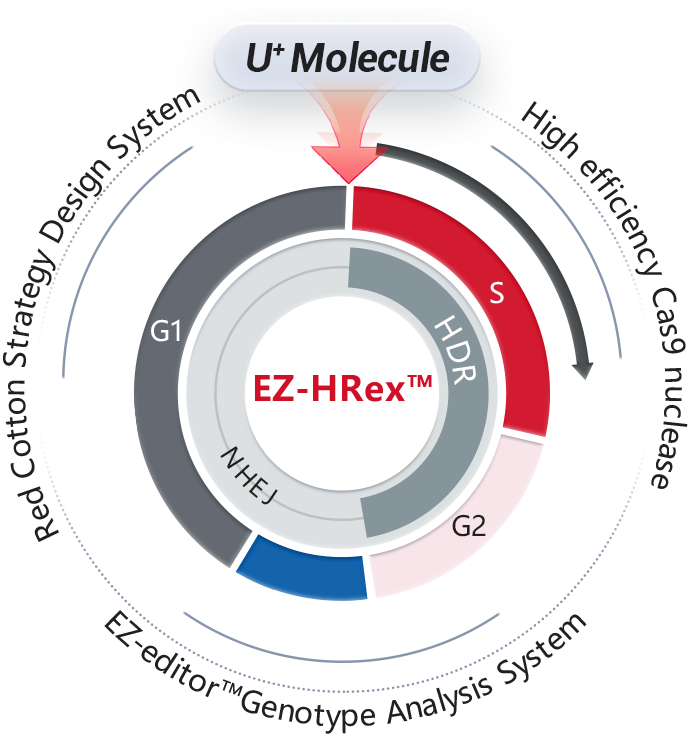
Stem CellIncubate sgRNA and Cas9 protein in vitro to form an RNP (ribonucleoprotein) complex, which is then co-transfected into cells along with a single-stranded oligonucleotide. After the RNP induces a double-strand break at the target site, the oligonucleotide can introduce the desired mutation into the genomic target through homologous recombination.
Features:
Applicable Types:
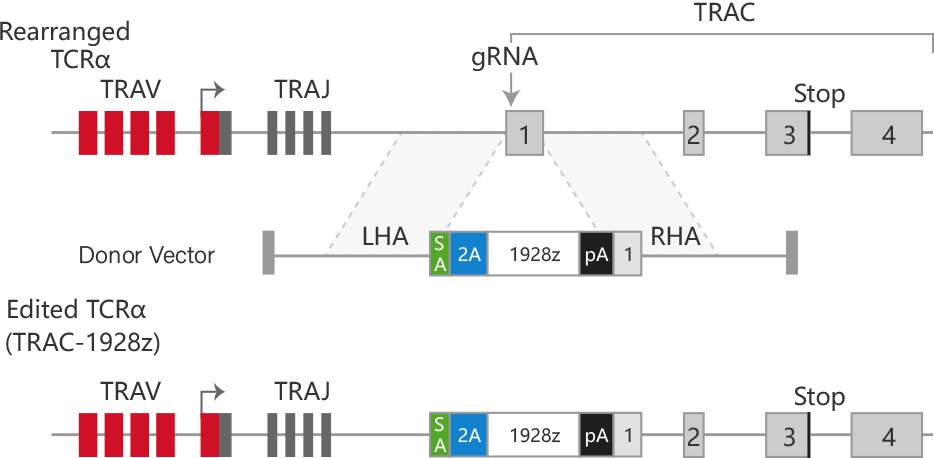
Gene editing is achieved by fusing an engineered reverse transcriptase with a Cas9 nickase and using a special prime editing guide RNA (pegRNA).
Features:
Applicable Types:
Incubate sgRNA and Cas9 protein in vitro to form an RNP (ribonucleoprotein) complex, which is then co-transfected into cells along with a single-stranded oligonucleotide. After the RNP induces a double-strand break at the target site, the oligonucleotide can introduce the desired mutation into the genomic target through homologous recombination.
Features:
Applicable Types:
Gene editing is achieved by fusing an engineered reverse transcriptase with a Cas9 nickase and using a special prime editing guide RNA (pegRNA).
Features:
Applicable Types: