

Location:Home > Application > Understanding on Common Cell Models for Colorectal Cancer Research
Understanding on Common Cell Models for Colorectal Cancer Research
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide. Colonoscopy screening helps with the early detection of CRC, but more than 25% of patients have metastases at diagnosis. Cancer metastasis is the leading cause of death among CRC patients, with a 5-year overall survival rate of less than 20%. CRC is a heterogeneous group of tumors that can be subdivided based on molecular characteristics to implement different treatment strategies. For example, Microsatellite Instability (MSI) and Microsatellite Stability (MSS), and the molecular profiling of metastatic CRC (including the analysis of gene mutations such as KRAS, NRAS, BRAF, PIK3CA) are increasingly important for determining prognosis and predictive biomarkers, as well as understanding the biological drivers of tumors. Clinical trials are currently selecting specific tumor features and using new drugs to target these tumors. Cell line models are widely used for drug sensitivity testing and identifying potential therapeutic compounds for patient treatment. This article introduces several common CRC cell lines, summarizes their molecular characteristics[2], and lists their applications in gene editing.
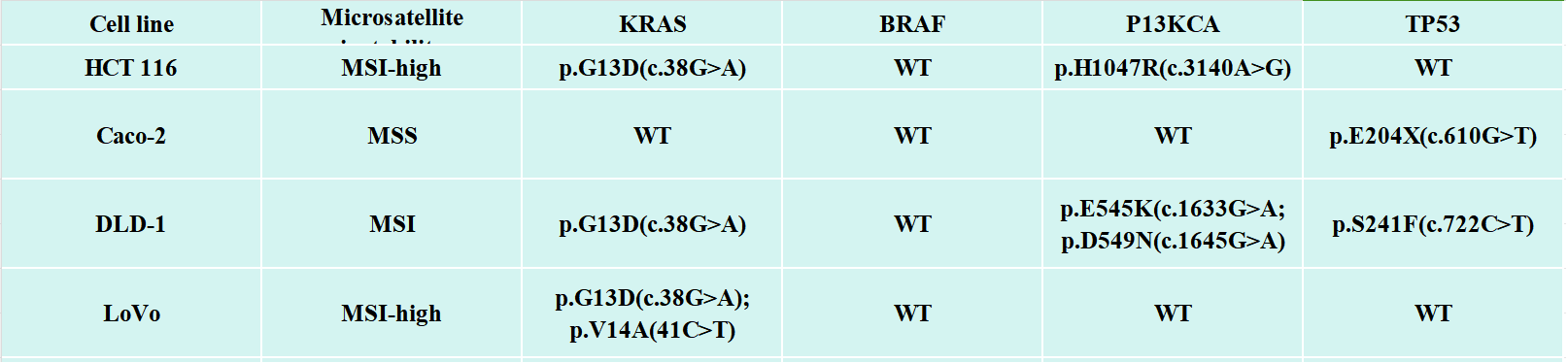
Figure 1. Molecular characteristics of different CRC cell lines
Catalog# YC-C004
Origin and Characteristics: HCT 116 is one of three cell lines established in 1979 by M. Brattain et al. from a male patient with colon cancer. HCT 116 forms clones in semi-solid agar and forms epithelial-like tumors in immunodeficient mice. This cell line has active cell proliferation, high clonogenic ability, drug resistance, as well as invasive and metastatic capabilities, making it widely used for studies on the pathogenesis of colon cancer, drug screening, therapy, and related signaling pathways.
Application Case: HCT 116 is a stable diploid human cell line commonly used as a tool cell, very suitable for large-scale gene screening. During the translation of the SARS-CoV-2 RNA, the host ribosome undergoes a programmed ribosomal frameshift (PRF) at a conserved structural element, which is essential for the replication of coronaviruses. However, the host factors regulating this process have not yet been identified. To identify the regulatory factors of SARS-CoV-2 PRF, researchers used HCT 116 cells to establish areporter cell clone expressing PRF-1 and PRF0, and conducted a genome-wide CRISPR-Cas9 knockout screening. The screening results indicated that the absence of ribosome recycling factors significantly reduced PRF efficiency and impaired the replication of the SARS-CoV-2 virus.The study revealed host factors that support efficient translation and replication of SARS-CoV-2[4].
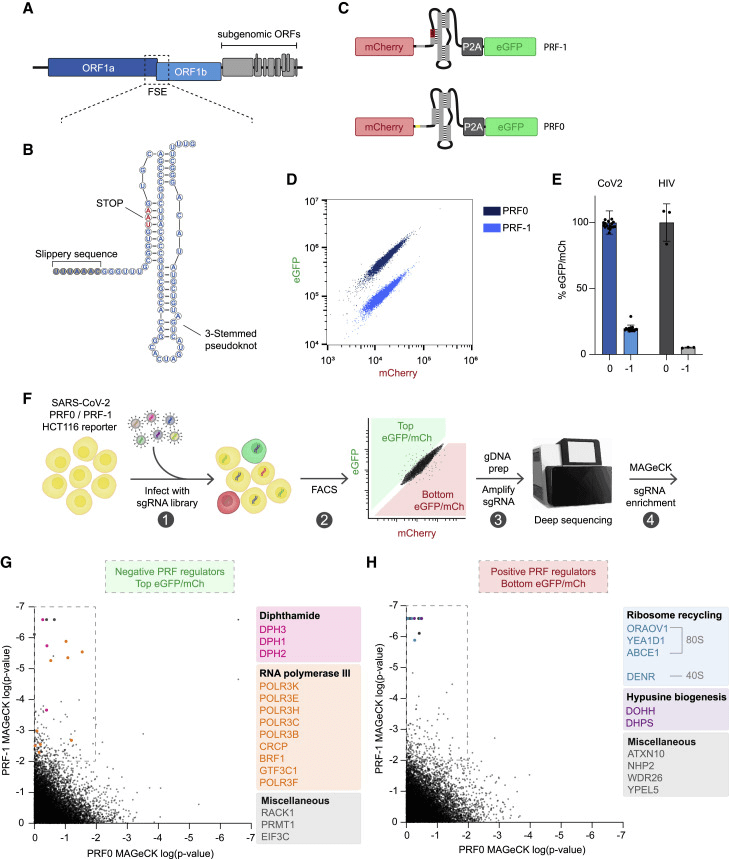
Figure 2. Identification of regulatory factors for SARS-CoV-2 PRF through whole-genome CRISPR-Cas9 knockout screening.
Discover More In-stock Cells >>>
Origin and Characteristics: The DLD-1 cell line, derived from human colorectal adenocarcinoma epithelial cells, is one of the two colorectal adenocarcinoma cell lines isolated by D.L. Dexter and colleagues between 1977 and 1979. These cells can form tumors in nude mice and are widely used in tumor research.
Application Case:
Due to a lack of efficacy or safety concerns with various Wnt signaling inhibitors, the clinical success rate is limited, driving people to find new targets in the Wnt signaling pathway. Researchers found that OBD9 has broad anti-cancer activity, particularly as an effective oral drug against colorectal cancer cells. To determine the mechanism, the researchers used HCT 116 and DLD-1 cells dealing with the drug and found that OBD9 exhibited cell viability-inhibiting activity similar to that of the potent TNIK kinase inhibitor NCB846. To investigate whether the inhibitory effect of OBD9 on cell viability might be due to TNIK suppression, researchers constructed TNIK inducible knockout cells (HCT116 iCas9–sgTNIK and DLD-1 iCas9-sgTNIK) and observed a decrease in cell viability following TNIK knockout. Additionally, HCT116 xenograft models were used to further confirm the inhibitory effect of OBD9 on the growth of colorectal cancer cells in vivo. The results indicated that this inhibitory effect is partly due to the autophagic mechanism in colorectal cancer cells promoting the degradation of TNIK, representing a new anti-tumor approach[3].
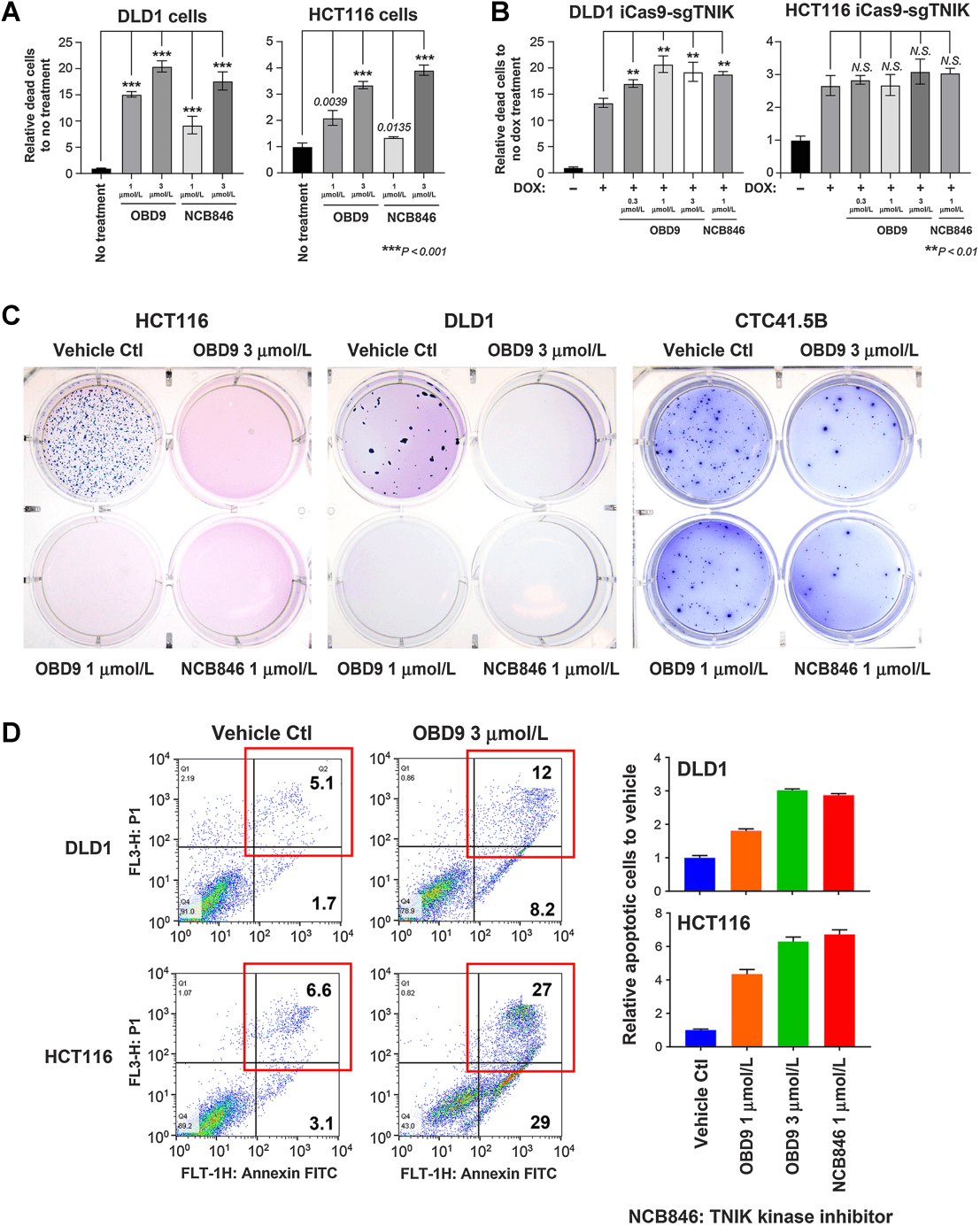
Figure 3. OBD9 inhibits the growth of colon cancer cells in vitro.
Catalog# YC-D003
Origin and Characteristics: Caco-2 cells were isolated from a 72-year-old male patient with rectal cancer. This cell line serves as a suitable transfection host and has value in tumor and toxicological research. When fully confluent, the cell lines exhibit characteristics of intestinal epithelial cell differentiation.
Application Case:
Long non-coding RNAs (lncRNAs) have been shown to be involved in various biological processes and confer resistance. However, it is still unclear whether lncRNAs are involved in the resistance effect of cetuximab in colorectal cancer cells. Researchers utilized Caco-2 cells to establish cetuximab-resistant cells (Caco-2 CR). It was found that lncRNA CRART16 was upregulated in Caco-2 CR cells, and then researchers constructed the Caco-2 cells with stable expression of CRART16 using the lentivirus method.The overexpression of CRART16 reversed the impact of cetuximab on cell viability and reduced cetuximab-induced cell apoptosis. Additionally, the overexpression of CRART16 resulted in an increase in the proportion of CD44+/CD133+ cells. Furthermore, CRART16 acts as a competitive endogenous RNA (ceRNA) for miR-371a-5p, which can regulate the expression of ERBB3. The miR-371a-5p mimic counteracted the cetuximab resistance induced by CRART16 overexpression. KEGG analysis indicated that the differentially expressed mRNAs generated upon CRART16 overexpression were primarily enriched in the MAPK signaling pathway[5].
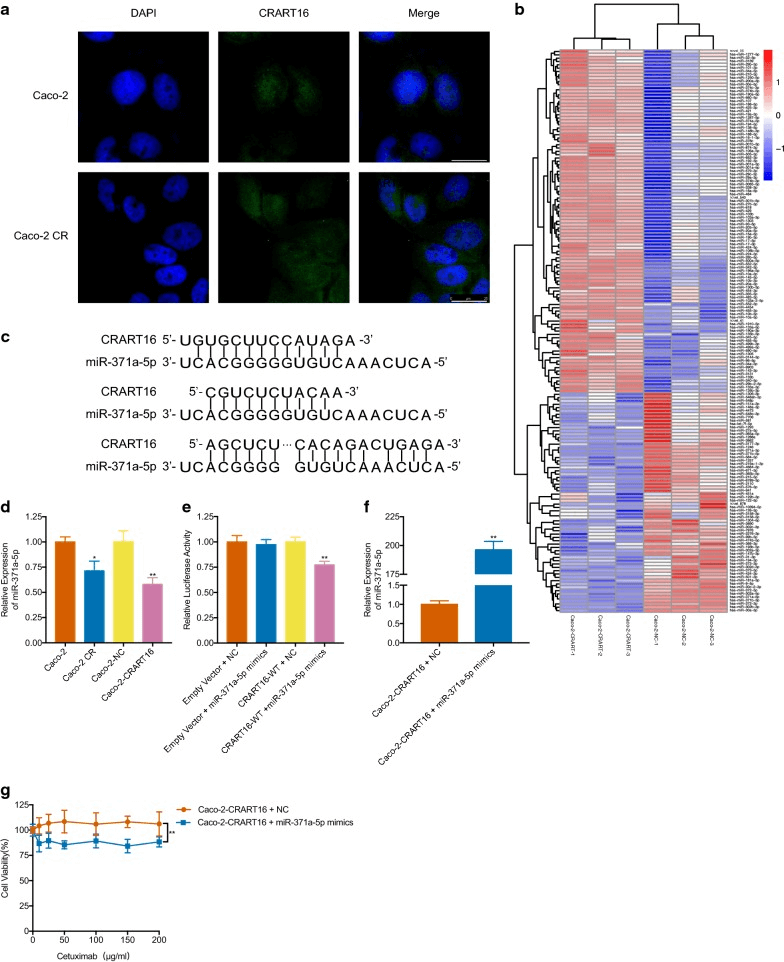
Figure 4. CRART16 promotes cetuximab resistance by downregulating the expression of miR-371a-5p.
Catalog# YC-C137
Origin and Characteristics: LoVo cells are derived from a tumor nodule from a 56-year-old male diagnosed with colon adenocarcinoma (with metastasis). They are utilized in cancer, toxicology, and immuno-oncology research, and can form tumors in nude mice.
Application Case:
PRL-3 plays a pathogenic role in tumor metastasis, but its underlying mechanisms are not yet fully understood. Initially, it was discovered that PRL-3 is upregulated in liver metastatic foci of colorectal cancer, while showing low or no expression in normal colorectal epithelium, adenomas, and primary lesions. Based on observations from previous studies indicating that PRL-3 can decrease the tyrosine phosphorylation of integrin β1 in HEK293 cells and enhance the activation of ERK1/2, researchers explored the association between PRL-3 and integrin β1 signaling, as well as its functional significance in the movement, invasion, and metastasis of colon cancer cells LoVo. By constructing LoVo cells stably expressing PRL-3 (LoVo-P), the enhanced phosphorylation of ERK1/2 was validated. Furthermore, transient transduction of integrin β1 siRNA in vitro revealed that integrin β1 is essential for PRL-3-induced cell movement and invasion. Additionally, shRNA-integrin β1 and shRNA-PRL-3 interference cell lines were constructed to study the requirement of integrin β1 for PRL-3-mediated metastasis in nude BALB/c mice. It was found that integrin β1 not only inhibits PRL-3-induced ERK1/2 phosphorylation but also reduces PRL-3-mediated cell movement, invasion, and lung metastasis in nude mice. Downstream of the integrin β1 pathway, ERK1/2 phosphorylation and MMP2 activity were identified as the reasons for PRL-3-mediated cell invasion. In summary, the study indicates that the integrin β1-ERK1/2-MMP2 signaling pathway plays a critical role in PRL-3-promoted movement, invasion, and metastasis of colorectal cancer cells[6].
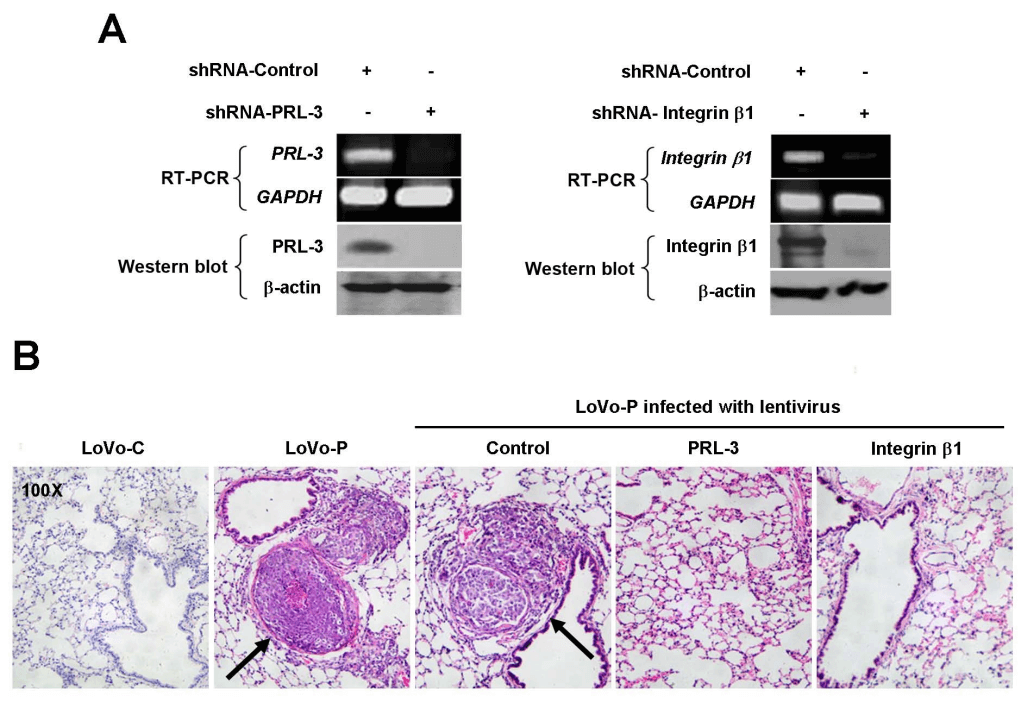
Figure 5. Depletion of integrin β1 or PRL-3 eliminates the promoting effect of PRL-3 on metastasis in vivo.
The above-mentioned commonly used colorectal cancer (CRC) cell models play an indispensable role in CRC research due to their unique molecular characteristics and biological behaviors. Through these cell lines, researchers can simulate the pathological processes of CRC and test drug sensitivity, which not only deepens our understanding of CRC but also provides valuable tools for the development of new therapeutic strategies.
Ubigenr offers gene-editing (KO/KI/PM) and stable cell line custom services related to colorectal cancer research—please feel free to inquire!
References
[1] Ciombor KK, Wu C, Goldberg RM. Recent therapeutic advances in the treatment of colorectal cancer. Annu Rev Med. 2015;66:83-95. doi: 10.1146/annurev-med-051513-102539.
[2] Berg KCG, Eide PW, Eilertsen IA, Johannessen B, Bruun J, Danielsen SA, Bjørnslett M, Meza-Zepeda LA, Eknæs M, Lind GE, Myklebost O, Skotheim RI, Sveen A, Lothe RA. Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Mol Cancer. 2017 Jul 6;16(1):116. doi: 10.1186/s12943-017-0691-y.
[3] Zhou K, Cheong JE, Krishnaji ST, Ghalali A, Fu H, Sui L, Alix-Panabières C, Cayrefourcq L, Bielenberg D, Sun L, Zetter B. Inhibition of Wnt Signaling in Colon Cancer Cells via an Oral Drug that Facilitates TNIK Degradation. Mol Cancer Ther. 2023 Jan 3;22(1):25-36. doi: 10.1158/1535-7163.MCT-21-0801.
[4] Rehfeld F, Eitson JL, Ohlson MB, Chang TC, Schoggins JW, Mendell JT. CRISPR screening reveals a dependency on ribosome recycling for efficient SARS-CoV-2 programmed ribosomal frameshifting and viral replication. Cell Rep. 2023 Feb 28;42(2):112076. doi: 10.1016/j.celrep.2023.112076.
[5] Zhang X, Wen L, Chen S, Zhang J, Ma Y, Hu J, Yue T, Wang J, Zhu J, Bu D, Wang X. The novel long noncoding RNA CRART16 confers cetuximab resistance in colorectal cancer cells by enhancing ERBB3 expression via miR-371a-5p. Cancer Cell Int. 2020 Mar 4;20:68. doi: 10.1186/s12935-020-1155-9.
[6] Peng L, Xing X, Li W, Qu L, Meng L, Lian S, Jiang B, Wu J, Shou C. PRL-3 promotes the motility, invasion, and metastasis of LoVo colon cancer cells through PRL-3-integrin beta1-ERK1/2 and-MMP2 signaling. Mol Cancer. 2009 Nov 24;8:110. doi: 10.1186/1476-4598-8-110.





Understanding on Common Cell Models for Colorectal Cancer Research

Colorectal cancer (CRC) is one of the most common malignant tumors worldwide. Colonoscopy screening helps with the early detection of CRC, but more than 25% of patients have metastases at diagnosis. Cancer metastasis is the leading cause of death among CRC patients, with a 5-year overall survival rate of less than 20%. CRC is a heterogeneous group of tumors that can be subdivided based on molecular characteristics to implement different treatment strategies. For example, Microsatellite Instability (MSI) and Microsatellite Stability (MSS), and the molecular profiling of metastatic CRC (including the analysis of gene mutations such as KRAS, NRAS, BRAF, PIK3CA) are increasingly important for determining prognosis and predictive biomarkers, as well as understanding the biological drivers of tumors. Clinical trials are currently selecting specific tumor features and using new drugs to target these tumors. Cell line models are widely used for drug sensitivity testing and identifying potential therapeutic compounds for patient treatment. This article introduces several common CRC cell lines, summarizes their molecular characteristics[2], and lists their applications in gene editing.

Figure 1. Molecular characteristics of different CRC cell lines
Catalog# YC-C004
Origin and Characteristics: HCT 116 is one of three cell lines established in 1979 by M. Brattain et al. from a male patient with colon cancer. HCT 116 forms clones in semi-solid agar and forms epithelial-like tumors in immunodeficient mice. This cell line has active cell proliferation, high clonogenic ability, drug resistance, as well as invasive and metastatic capabilities, making it widely used for studies on the pathogenesis of colon cancer, drug screening, therapy, and related signaling pathways.
Application Case: HCT 116 is a stable diploid human cell line commonly used as a tool cell, very suitable for large-scale gene screening. During the translation of the SARS-CoV-2 RNA, the host ribosome undergoes a programmed ribosomal frameshift (PRF) at a conserved structural element, which is essential for the replication of coronaviruses. However, the host factors regulating this process have not yet been identified. To identify the regulatory factors of SARS-CoV-2 PRF, researchers used HCT 116 cells to establish areporter cell clone expressing PRF-1 and PRF0, and conducted a genome-wide CRISPR-Cas9 knockout screening. The screening results indicated that the absence of ribosome recycling factors significantly reduced PRF efficiency and impaired the replication of the SARS-CoV-2 virus.The study revealed host factors that support efficient translation and replication of SARS-CoV-2[4].

Figure 2. Identification of regulatory factors for SARS-CoV-2 PRF through whole-genome CRISPR-Cas9 knockout screening.
Discover More In-stock Cells >>>
Origin and Characteristics: The DLD-1 cell line, derived from human colorectal adenocarcinoma epithelial cells, is one of the two colorectal adenocarcinoma cell lines isolated by D.L. Dexter and colleagues between 1977 and 1979. These cells can form tumors in nude mice and are widely used in tumor research.
Application Case:
Due to a lack of efficacy or safety concerns with various Wnt signaling inhibitors, the clinical success rate is limited, driving people to find new targets in the Wnt signaling pathway. Researchers found that OBD9 has broad anti-cancer activity, particularly as an effective oral drug against colorectal cancer cells. To determine the mechanism, the researchers used HCT 116 and DLD-1 cells dealing with the drug and found that OBD9 exhibited cell viability-inhibiting activity similar to that of the potent TNIK kinase inhibitor NCB846. To investigate whether the inhibitory effect of OBD9 on cell viability might be due to TNIK suppression, researchers constructed TNIK inducible knockout cells (HCT116 iCas9–sgTNIK and DLD-1 iCas9-sgTNIK) and observed a decrease in cell viability following TNIK knockout. Additionally, HCT116 xenograft models were used to further confirm the inhibitory effect of OBD9 on the growth of colorectal cancer cells in vivo. The results indicated that this inhibitory effect is partly due to the autophagic mechanism in colorectal cancer cells promoting the degradation of TNIK, representing a new anti-tumor approach[3].

Figure 3. OBD9 inhibits the growth of colon cancer cells in vitro.
Catalog# YC-D003
Origin and Characteristics: Caco-2 cells were isolated from a 72-year-old male patient with rectal cancer. This cell line serves as a suitable transfection host and has value in tumor and toxicological research. When fully confluent, the cell lines exhibit characteristics of intestinal epithelial cell differentiation.
Application Case:
Long non-coding RNAs (lncRNAs) have been shown to be involved in various biological processes and confer resistance. However, it is still unclear whether lncRNAs are involved in the resistance effect of cetuximab in colorectal cancer cells. Researchers utilized Caco-2 cells to establish cetuximab-resistant cells (Caco-2 CR). It was found that lncRNA CRART16 was upregulated in Caco-2 CR cells, and then researchers constructed the Caco-2 cells with stable expression of CRART16 using the lentivirus method.The overexpression of CRART16 reversed the impact of cetuximab on cell viability and reduced cetuximab-induced cell apoptosis. Additionally, the overexpression of CRART16 resulted in an increase in the proportion of CD44+/CD133+ cells. Furthermore, CRART16 acts as a competitive endogenous RNA (ceRNA) for miR-371a-5p, which can regulate the expression of ERBB3. The miR-371a-5p mimic counteracted the cetuximab resistance induced by CRART16 overexpression. KEGG analysis indicated that the differentially expressed mRNAs generated upon CRART16 overexpression were primarily enriched in the MAPK signaling pathway[5].

Figure 4. CRART16 promotes cetuximab resistance by downregulating the expression of miR-371a-5p.
Catalog# YC-C137
Origin and Characteristics: LoVo cells are derived from a tumor nodule from a 56-year-old male diagnosed with colon adenocarcinoma (with metastasis). They are utilized in cancer, toxicology, and immuno-oncology research, and can form tumors in nude mice.
Application Case:
PRL-3 plays a pathogenic role in tumor metastasis, but its underlying mechanisms are not yet fully understood. Initially, it was discovered that PRL-3 is upregulated in liver metastatic foci of colorectal cancer, while showing low or no expression in normal colorectal epithelium, adenomas, and primary lesions. Based on observations from previous studies indicating that PRL-3 can decrease the tyrosine phosphorylation of integrin β1 in HEK293 cells and enhance the activation of ERK1/2, researchers explored the association between PRL-3 and integrin β1 signaling, as well as its functional significance in the movement, invasion, and metastasis of colon cancer cells LoVo. By constructing LoVo cells stably expressing PRL-3 (LoVo-P), the enhanced phosphorylation of ERK1/2 was validated. Furthermore, transient transduction of integrin β1 siRNA in vitro revealed that integrin β1 is essential for PRL-3-induced cell movement and invasion. Additionally, shRNA-integrin β1 and shRNA-PRL-3 interference cell lines were constructed to study the requirement of integrin β1 for PRL-3-mediated metastasis in nude BALB/c mice. It was found that integrin β1 not only inhibits PRL-3-induced ERK1/2 phosphorylation but also reduces PRL-3-mediated cell movement, invasion, and lung metastasis in nude mice. Downstream of the integrin β1 pathway, ERK1/2 phosphorylation and MMP2 activity were identified as the reasons for PRL-3-mediated cell invasion. In summary, the study indicates that the integrin β1-ERK1/2-MMP2 signaling pathway plays a critical role in PRL-3-promoted movement, invasion, and metastasis of colorectal cancer cells[6].

Figure 5. Depletion of integrin β1 or PRL-3 eliminates the promoting effect of PRL-3 on metastasis in vivo.
The above-mentioned commonly used colorectal cancer (CRC) cell models play an indispensable role in CRC research due to their unique molecular characteristics and biological behaviors. Through these cell lines, researchers can simulate the pathological processes of CRC and test drug sensitivity, which not only deepens our understanding of CRC but also provides valuable tools for the development of new therapeutic strategies.
Ubigenr offers gene-editing (KO/KI/PM) and stable cell line custom services related to colorectal cancer research—please feel free to inquire!
References
[1] Ciombor KK, Wu C, Goldberg RM. Recent therapeutic advances in the treatment of colorectal cancer. Annu Rev Med. 2015;66:83-95. doi: 10.1146/annurev-med-051513-102539.
[2] Berg KCG, Eide PW, Eilertsen IA, Johannessen B, Bruun J, Danielsen SA, Bjørnslett M, Meza-Zepeda LA, Eknæs M, Lind GE, Myklebost O, Skotheim RI, Sveen A, Lothe RA. Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Mol Cancer. 2017 Jul 6;16(1):116. doi: 10.1186/s12943-017-0691-y.
[3] Zhou K, Cheong JE, Krishnaji ST, Ghalali A, Fu H, Sui L, Alix-Panabières C, Cayrefourcq L, Bielenberg D, Sun L, Zetter B. Inhibition of Wnt Signaling in Colon Cancer Cells via an Oral Drug that Facilitates TNIK Degradation. Mol Cancer Ther. 2023 Jan 3;22(1):25-36. doi: 10.1158/1535-7163.MCT-21-0801.
[4] Rehfeld F, Eitson JL, Ohlson MB, Chang TC, Schoggins JW, Mendell JT. CRISPR screening reveals a dependency on ribosome recycling for efficient SARS-CoV-2 programmed ribosomal frameshifting and viral replication. Cell Rep. 2023 Feb 28;42(2):112076. doi: 10.1016/j.celrep.2023.112076.
[5] Zhang X, Wen L, Chen S, Zhang J, Ma Y, Hu J, Yue T, Wang J, Zhu J, Bu D, Wang X. The novel long noncoding RNA CRART16 confers cetuximab resistance in colorectal cancer cells by enhancing ERBB3 expression via miR-371a-5p. Cancer Cell Int. 2020 Mar 4;20:68. doi: 10.1186/s12935-020-1155-9.
[6] Peng L, Xing X, Li W, Qu L, Meng L, Lian S, Jiang B, Wu J, Shou C. PRL-3 promotes the motility, invasion, and metastasis of LoVo colon cancer cells through PRL-3-integrin beta1-ERK1/2 and-MMP2 signaling. Mol Cancer. 2009 Nov 24;8:110. doi: 10.1186/1476-4598-8-110.






