

Location:Home > Application > Human Metabolic Gene Knockout Library Makes a Big Impact in Tumor Research
Human Metabolic Gene Knockout Library Makes a Big Impact in Tumor Research

In the field of tumor therapy, precision targeted treatment is the key point, as it targets cancer-specific molecules and reduces damage to normal cells. To achieve this, it is essential to gain an in-depth understanding of the metabolic pathways in cancer cells and the function of the associated genes. Today, Ubigene introduces a powerful tool——the Human Metabolic Gene Knockout Library, which targets about 3000 metabolic enzymes, small molecule transporters, and metabolism-related transcription factors in the human genome, providing strong support for relevant research. Let's explore the value and function of the human metabolic gene knockout library in cancer research through a case study.
Aurora kinase A (AURKA) is an important mitotic kinase, and its inhibitors, such as MLN8237, have shown antitumor effects across various cancer types. However, improving the efficacy and selectivity of these inhibitors remains a challenge. Additionally, the KEAP1-NRF2 signaling pathway plays a central role in tumor metabolism, particularly in non-small cell lung cancer (NSCLC) with KEAP1 mutations or deficiencies, where these tumors exhibit high resistance to conventional therapies. Therefore, exploring new indications for AURKA inhibitors or their combination with other drugs has become a key direction for enhancing efficacy. This research aims to identify specific tumor subtypes or metabolic characteristics that are sensitive to AURKA inhibitors, providing new strategies for clinical treatment.
In this study, to identify metabolic indications for AURKA inhibitors, researchers used CRISPR/Cas9 technology to perform metabolic library screening in lung cancer and breast cancer cell lines.
Library Type: Human Metabolic Gene Knockout Library
Transduced Cells: MDA-MB-231 and H1975
Screening Method: The MDA-MB-231 and H1975 library cells were divided into DMSO plasmid group and the MLN8237 treatment groups (various concentrations). Cells were collected at day 3 and day 7 after treatment to ensure each group had over 2×10^7 cells for a 300X coverage. Genomic DNA was extracted, sgRNA constructs were amplified and sequenced, and sequencing was performed using the Illumina HiSeq2000 platform. Data analysis was carried out with the MAGeCK-MLE software.
The researchers found that the KEAP1 gene showed significant negative selection under the action of the AURKA inhibitor MLN8237. By analyzing the GDSC database, they confirmed that KEAP1-mutated NSCLC cells were more sensitive to MLN8237.
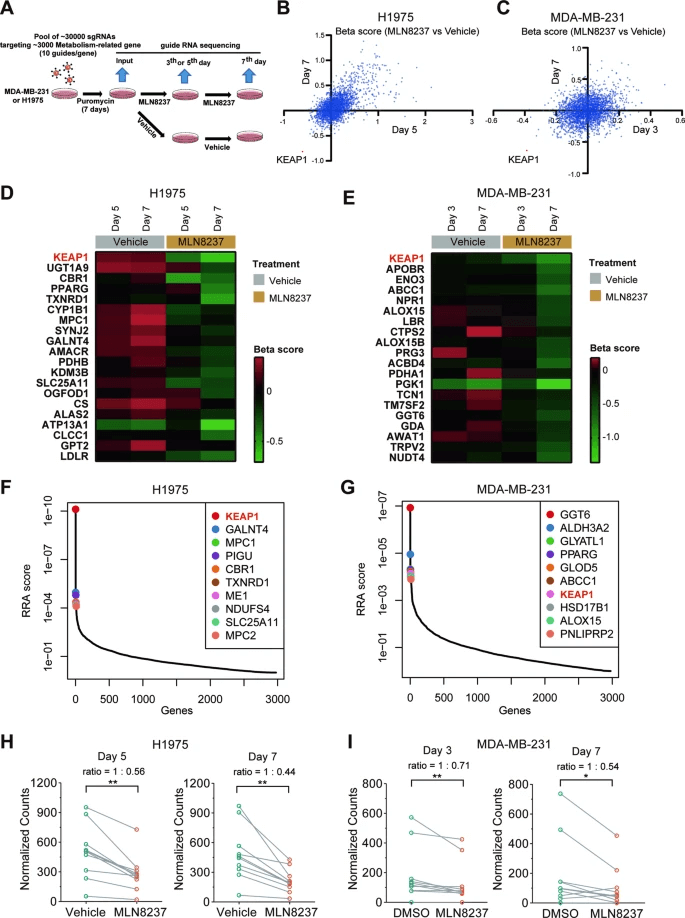
Figure 1: KEAP1 Is a Significant Negative Selection Gene for AURKA Inhibition.
Further CCK8 assays and colony formation experiments also supported this finding, showing that the loss of KEAP1 increased cell sensitivity to AURKA inhibitors. In nude mouse models, NSCLC cells with KEAP1 knockdown also exhibited increased sensitivity to MLN8237. Additionally, the study found that activation of the NRF2 pathway played a key role in the sensitizing effect caused by KEAP1 knockdown, as knocking down NRF2 alleviated this increased sensitivity. This study revealed that KEAP1 is a key metabolic gene that affects sensitivity to AURKA inhibitors like MLN8237, providing a new target for future therapeutic strategies.
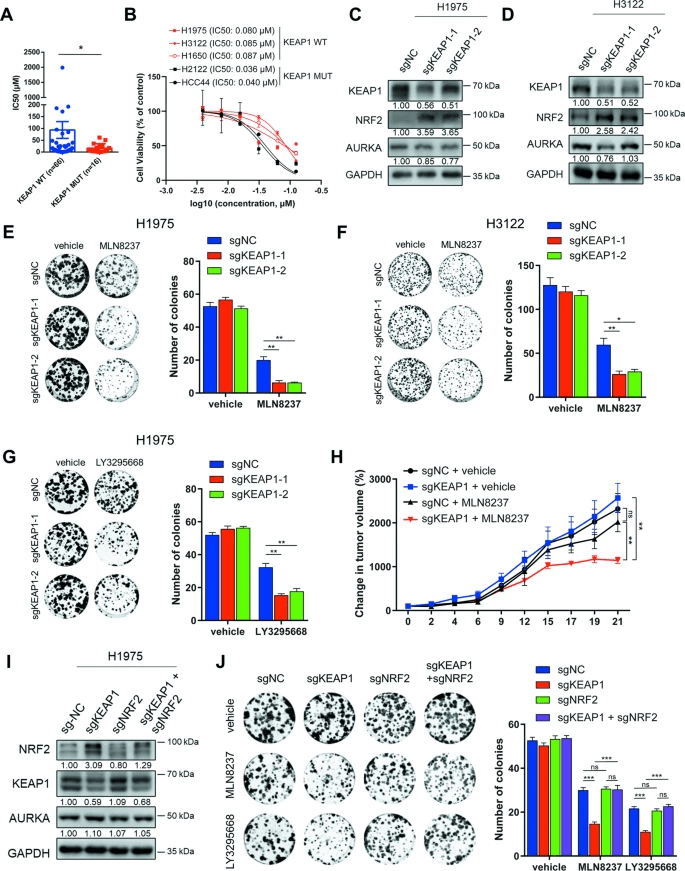
Figure 2: KEAP1 Deficiency Activates NRF2 Making NSCLC Cells Sensitive to AURKA Inhibition
Previous research has identified certain gene mutations related to tumor sensitivity to AURKA inhibitors, such as defects in RB1 and deletions in SMARCA4 or ARID1A. However, due to the differences in mutation patterns among different tumors, in order to comprehensively explore the application scope of AURKA inhibitors, it is necessary to use CRISPR/Cas9 technology for extensive screening in multiple tumor types. This study not only expands our understanding of tumor sensitivity to AURKA inhibitors but also highlights the importance of further in-depth exploration of different tumor types in future research.
The human metabolic gene knockout library has played a significant role in the aforementioned study, helping to uncover the metabolic mechanisms of drug resistance and identify new metabolic targets to overcome resistance. This provides a new avenue for combination therapy.
Ubigene offers human metabolic gene knockout library plasmids and viruses, screening-ready cell pools, also one-stop functional screening as low as $8K, delivering NGS analysis report and data.
For more information, feel free to send an inquiry!
References
Deng, B., Liu, F., Chen, N. et al. AURKA emerges as a vulnerable target for KEAP1-deficient non-small cell lung cancer by activation of asparagine synthesis. Cell Death Dis 15, 233 (2024). https://doi.org/10.1038/s41419-024-06577-x




Human Metabolic Gene Knockout Library Makes a Big Impact in Tumor Research

In the field of tumor therapy, precision targeted treatment is the key point, as it targets cancer-specific molecules and reduces damage to normal cells. To achieve this, it is essential to gain an in-depth understanding of the metabolic pathways in cancer cells and the function of the associated genes. Today, Ubigene introduces a powerful tool——the Human Metabolic Gene Knockout Library, which targets about 3000 metabolic enzymes, small molecule transporters, and metabolism-related transcription factors in the human genome, providing strong support for relevant research. Let's explore the value and function of the human metabolic gene knockout library in cancer research through a case study.
Aurora kinase A (AURKA) is an important mitotic kinase, and its inhibitors, such as MLN8237, have shown antitumor effects across various cancer types. However, improving the efficacy and selectivity of these inhibitors remains a challenge. Additionally, the KEAP1-NRF2 signaling pathway plays a central role in tumor metabolism, particularly in non-small cell lung cancer (NSCLC) with KEAP1 mutations or deficiencies, where these tumors exhibit high resistance to conventional therapies. Therefore, exploring new indications for AURKA inhibitors or their combination with other drugs has become a key direction for enhancing efficacy. This research aims to identify specific tumor subtypes or metabolic characteristics that are sensitive to AURKA inhibitors, providing new strategies for clinical treatment.
In this study, to identify metabolic indications for AURKA inhibitors, researchers used CRISPR/Cas9 technology to perform metabolic library screening in lung cancer and breast cancer cell lines.
Library Type: Human Metabolic Gene Knockout Library
Transduced Cells: MDA-MB-231 and H1975
Screening Method: The MDA-MB-231 and H1975 library cells were divided into DMSO plasmid group and the MLN8237 treatment groups (various concentrations). Cells were collected at day 3 and day 7 after treatment to ensure each group had over 2×10^7 cells for a 300X coverage. Genomic DNA was extracted, sgRNA constructs were amplified and sequenced, and sequencing was performed using the Illumina HiSeq2000 platform. Data analysis was carried out with the MAGeCK-MLE software.
The researchers found that the KEAP1 gene showed significant negative selection under the action of the AURKA inhibitor MLN8237. By analyzing the GDSC database, they confirmed that KEAP1-mutated NSCLC cells were more sensitive to MLN8237.

Figure 1: KEAP1 Is a Significant Negative Selection Gene for AURKA Inhibition.
Further CCK8 assays and colony formation experiments also supported this finding, showing that the loss of KEAP1 increased cell sensitivity to AURKA inhibitors. In nude mouse models, NSCLC cells with KEAP1 knockdown also exhibited increased sensitivity to MLN8237. Additionally, the study found that activation of the NRF2 pathway played a key role in the sensitizing effect caused by KEAP1 knockdown, as knocking down NRF2 alleviated this increased sensitivity. This study revealed that KEAP1 is a key metabolic gene that affects sensitivity to AURKA inhibitors like MLN8237, providing a new target for future therapeutic strategies.

Figure 2: KEAP1 Deficiency Activates NRF2 Making NSCLC Cells Sensitive to AURKA Inhibition
Previous research has identified certain gene mutations related to tumor sensitivity to AURKA inhibitors, such as defects in RB1 and deletions in SMARCA4 or ARID1A. However, due to the differences in mutation patterns among different tumors, in order to comprehensively explore the application scope of AURKA inhibitors, it is necessary to use CRISPR/Cas9 technology for extensive screening in multiple tumor types. This study not only expands our understanding of tumor sensitivity to AURKA inhibitors but also highlights the importance of further in-depth exploration of different tumor types in future research.
The human metabolic gene knockout library has played a significant role in the aforementioned study, helping to uncover the metabolic mechanisms of drug resistance and identify new metabolic targets to overcome resistance. This provides a new avenue for combination therapy.
Ubigene offers human metabolic gene knockout library plasmids and viruses, screening-ready cell pools, also one-stop functional screening as low as $8K, delivering NGS analysis report and data.
For more information, feel free to send an inquiry!
References
Deng, B., Liu, F., Chen, N. et al. AURKA emerges as a vulnerable target for KEAP1-deficient non-small cell lung cancer by activation of asparagine synthesis. Cell Death Dis 15, 233 (2024). https://doi.org/10.1038/s41419-024-06577-x




