Location:Home > Application > Discovery of Potential Clinical Targets for Gliomas Using CRISPR Libraries
Discovery of Potential Clinical Targets for Gliomas Using CRISPR Libraries

In the field of complex biological mechanisms of brain tumors, CRISPR libraries serve as essential tools to help researchers identify potential targets. This article illustrates the application of CRISPR libraries in studying high-grade gliomas, providing insights into how these libraries can facilitate breakthrough advances in brain tumor research.
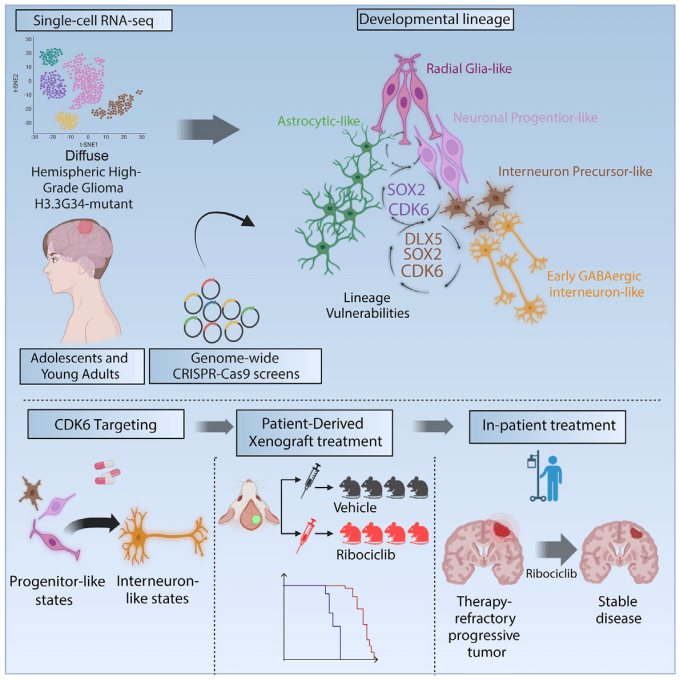
High-grade gliomas (HGG) are deadly primary brain cancers and are among the leading causes of cancer-related deaths in children and adolescents. Among these, the diffuse hemispheric gliomas with H3G34 mutations (DHG-H3G34) primarily occur in adolescents and account for over 30% of childhood or adolescent HGGs, but they have been relatively under-researched.
Illon Liu et al. conducted whole-genome CRISPR-Cas9 screenings to identify genes critical for the growth and survival of DHG-H3G34 cells, ultimately discovering potential clinically actionable targets. To gain a deeper understanding of the gene dependencies and vulnerabilities of DHG-H3G34 tumor cells, the researchers performed two distinct whole-genome library screenings and data analysis.
Library Type: Human Genome-wide Knockout Library (GeCKOv2 library)
Transduced Cells: OPBG-GBM-001 (derived from H3.3G34R patient)
Screening Method: T0 samples were collected and stored at -80°C for subsequent processing. The remaining cells were cultured without antibiotics for 10 passages (30 days), with fresh media added every 2-3 days, and passaged weekly. Gene expression at different time points (T0 vs. T8, T0 vs. T10) was compared to ascertain gene necessity.
Library Type: Human Genome-wide Knockout Library (Avana library)
Transduced Cells: KNS42
Screening Method: Cells were passaged without selection for 21 days, maintaining a 500-fold coverage of each sgRNA. Genomic DNA was extracted from endpoint cell pellets, followed by PCR amplification and sequencing using standard Illumina protocols.
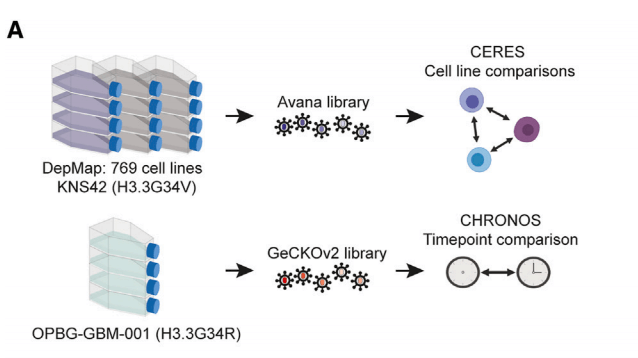
Figure 1: Schematic of whole-genome CRISPR-Cas9 screening.
When projecting the genes identified from the two CRISPR library screenings onto single-cell RNA sequencing dataset from DHG-H3G34 patients, most hit genes were significantly enriched in progenitor-like cells related to intermediate neuronal lineage development, with almost no hit genes associated with AC-like/MES-like cell programs. This indicates that DHG-H3G34 primarily relies on gene characteristics related to the intermediate neuronal developmental state. In both screenings, SOX2 and CDK6 emerged as the most important hit genes based on ranked gene effect scores, showing high selectivity in 769 pan-cancer cell lines assessed by DepMap consortium.
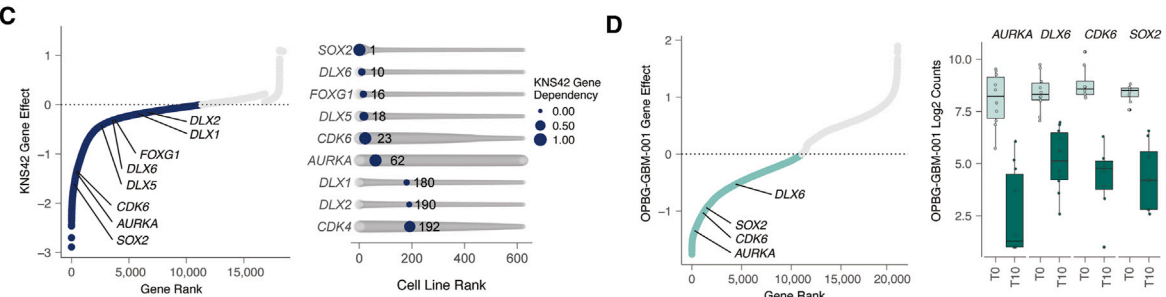
Figure 2: Gene ranking from KNS42 screening (C); Gene ranking from OPBG-GBM-001 screening (D).
Functional Validation of CDK6 Gene
To move the findings toward more direct clinical applicability, the researcher validated CDK6 as a target available for FDA-approved drugs, which was shown to be one of the most specific targets for DHG-H3G34 in comparison to 700+ pan-cancer cell lines. In vitro validation revealed that the knockout of CDK6 significantly reduced the viability of various DHG-H3G34 cells.
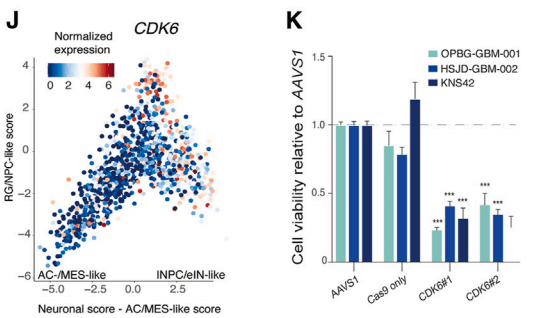
Figure 3: Significant decrease in cell viability after gene knockout screening.
The depletion of CDK6 was validated in vivo through two methods. After the knockout of CDK6, BT690-DHG-H3G34 tumor cells were xenotransplanted into mice; following the formation of tumors from the BT690 xenografts, shRNA-mediated CDK6 knockdown was induced. Both methods significantly inhibited tumor growth and enhanced overall survival.
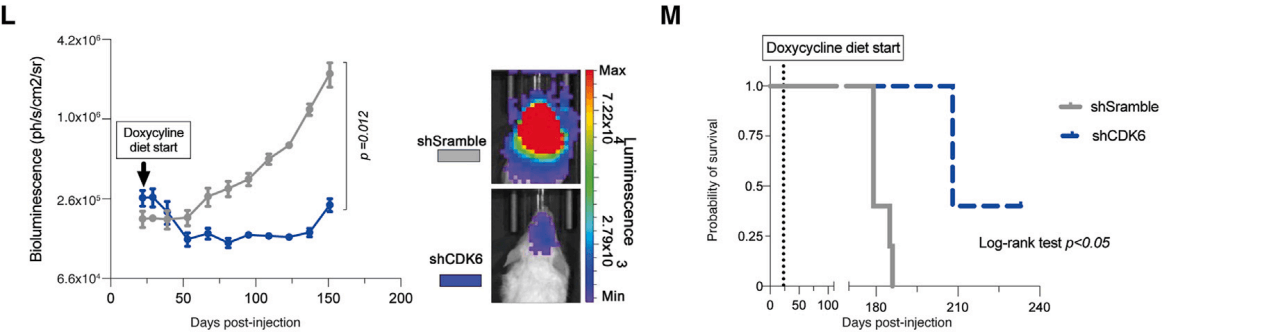
Figure 4: Growth changes in DHG-H3G34 tumors under gene knockout
Furthermore, CDK6 was found to be upregulated in tumor cells through epigenetic mechanisms. Its specific degraders and CDK4/6 inhibitors (such as Ribociclib) effectively reduced tumor growth and extended the survival of xenotransplanted mice. In a clinical case, a DHG-H3G34 patient stabilized for 18 cycles after treatment with Ribociclib.
This study reveals that DHG-H3G34 recapitulates various stages of intermediate neuronal lineage development along a presumed developmental hierarchy and derives and validates unique targets from it. By integrating whole-genome CRISPR-Cas9 screening with single-cell RNA sequencing results, it demonstrates that DHG-H3G34 depends on genes from progenitor-like compartments in the intermediate neuronal lineage hierarchy, and targeting CDK6 can modify its characteristic developmental biology, paving the way for further translational research.
It is evident that the application of CRISPR library screening techniques in the field of brain tumor research not only provides researchers with a powerful gene editing tool but also offers valuable resources for understanding tumor complexities, discovering new therapeutic targets, advancing personalized medicine, and accelerating translational research.
Ubigene can provide the same library cited in this paper
[Human Genome-wide Knockout Library] Plasmids | Virus | Screening-Ready Cell Pool
Along with one-stop functional screening from $8K only
Delivering NGS analysis report and data
Inquire now to design your own library cell screening project
Reference
Liu, Ilon et al. “GABAergic neuronal lineage development determines clinically actionable targets in diffuse hemispheric glioma, H3G34-mutant.” Cancer cell, S1535-6108(24)00305-2. 27 Aug. 2024, doi:10.1016/j.ccell.2024.08.006


Discovery of Potential Clinical Targets for Gliomas Using CRISPR Libraries

In the field of complex biological mechanisms of brain tumors, CRISPR libraries serve as essential tools to help researchers identify potential targets. This article illustrates the application of CRISPR libraries in studying high-grade gliomas, providing insights into how these libraries can facilitate breakthrough advances in brain tumor research.
High-grade gliomas (HGG) are deadly primary brain cancers and are among the leading causes of cancer-related deaths in children and adolescents. Among these, the diffuse hemispheric gliomas with H3G34 mutations (DHG-H3G34) primarily occur in adolescents and account for over 30% of childhood or adolescent HGGs, but they have been relatively under-researched.
Illon Liu et al. conducted whole-genome CRISPR-Cas9 screenings to identify genes critical for the growth and survival of DHG-H3G34 cells, ultimately discovering potential clinically actionable targets. To gain a deeper understanding of the gene dependencies and vulnerabilities of DHG-H3G34 tumor cells, the researchers performed two distinct whole-genome library screenings and data analysis.

Library Type: Human Genome-wide Knockout Library (GeCKOv2 library)
Transduced Cells: OPBG-GBM-001 (derived from H3.3G34R patient)
Screening Method: T0 samples were collected and stored at -80°C for subsequent processing. The remaining cells were cultured without antibiotics for 10 passages (30 days), with fresh media added every 2-3 days, and passaged weekly. Gene expression at different time points (T0 vs. T8, T0 vs. T10) was compared to ascertain gene necessity.
Library Type: Human Genome-wide Knockout Library (Avana library)
Transduced Cells: KNS42
Screening Method: Cells were passaged without selection for 21 days, maintaining a 500-fold coverage of each sgRNA. Genomic DNA was extracted from endpoint cell pellets, followed by PCR amplification and sequencing using standard Illumina protocols.

Figure 1: Schematic of whole-genome CRISPR-Cas9 screening.
When projecting the genes identified from the two CRISPR library screenings onto single-cell RNA sequencing dataset from DHG-H3G34 patients, most hit genes were significantly enriched in progenitor-like cells related to intermediate neuronal lineage development, with almost no hit genes associated with AC-like/MES-like cell programs. This indicates that DHG-H3G34 primarily relies on gene characteristics related to the intermediate neuronal developmental state. In both screenings, SOX2 and CDK6 emerged as the most important hit genes based on ranked gene effect scores, showing high selectivity in 769 pan-cancer cell lines assessed by DepMap consortium.

Figure 2: Gene ranking from KNS42 screening (C); Gene ranking from OPBG-GBM-001 screening (D).
Functional Validation of CDK6 Gene
To move the findings toward more direct clinical applicability, the researcher validated CDK6 as a target available for FDA-approved drugs, which was shown to be one of the most specific targets for DHG-H3G34 in comparison to 700+ pan-cancer cell lines. In vitro validation revealed that the knockout of CDK6 significantly reduced the viability of various DHG-H3G34 cells.

Figure 3: Significant decrease in cell viability after gene knockout screening.
The depletion of CDK6 was validated in vivo through two methods. After the knockout of CDK6, BT690-DHG-H3G34 tumor cells were xenotransplanted into mice; following the formation of tumors from the BT690 xenografts, shRNA-mediated CDK6 knockdown was induced. Both methods significantly inhibited tumor growth and enhanced overall survival.

Figure 4: Growth changes in DHG-H3G34 tumors under gene knockout
Furthermore, CDK6 was found to be upregulated in tumor cells through epigenetic mechanisms. Its specific degraders and CDK4/6 inhibitors (such as Ribociclib) effectively reduced tumor growth and extended the survival of xenotransplanted mice. In a clinical case, a DHG-H3G34 patient stabilized for 18 cycles after treatment with Ribociclib.
This study reveals that DHG-H3G34 recapitulates various stages of intermediate neuronal lineage development along a presumed developmental hierarchy and derives and validates unique targets from it. By integrating whole-genome CRISPR-Cas9 screening with single-cell RNA sequencing results, it demonstrates that DHG-H3G34 depends on genes from progenitor-like compartments in the intermediate neuronal lineage hierarchy, and targeting CDK6 can modify its characteristic developmental biology, paving the way for further translational research.
It is evident that the application of CRISPR library screening techniques in the field of brain tumor research not only provides researchers with a powerful gene editing tool but also offers valuable resources for understanding tumor complexities, discovering new therapeutic targets, advancing personalized medicine, and accelerating translational research.
Ubigene can provide the same library cited in this paper
[Human Genome-wide Knockout Library] Plasmids | Virus | Screening-Ready Cell Pool
Along with one-stop functional screening from $8K only
Delivering NGS analysis report and data
Inquire now to design your own library cell screening project
Reference
Liu, Ilon et al. “GABAergic neuronal lineage development determines clinically actionable targets in diffuse hemispheric glioma, H3G34-mutant.” Cancer cell, S1535-6108(24)00305-2. 27 Aug. 2024, doi:10.1016/j.ccell.2024.08.006


