

Location:Home > Application > Expert Insights | Cell Models Commonly Used in Liver Cancer Research
Expert Insights | Cell Models Commonly Used in Liver Cancer Research
Liver cancer, or primary liver cancer, is a prevalent malignant tumor worldwide. This disease typically shows no obvious symptoms in the early stages and progresses rapidly, with most patients being diagnosed at an advanced stage. Symptoms of liver cancer may include pain in the liver area, fatigue, weight loss, jaundice, and ascites. Cell models of liver cancer are crucial for studying the occurrence, development, and biological characteristics of the disease, providing essential experimental platforms for drug screening, treatment evaluation, and new drug development. Today, we will share some commonly used cell models in liver cancer research and their application cases.
Catalog#: YC-C001
Source: The Hep G2 cell line is derived from a 15-year-old Caucasian male with hepatoblastoma.
Molecular Expression: Hep G2 cells retain many genetic expression features of normal liver cells and can express various liver-specific genes.
Growth Characteristics: Similar morphology to normal liver cells, adherent growth.
Applications: Widely used in various liver-related research areas, including drug toxicity assessments, bioartificial liver system construction, and studies on hepatitis B virus models.
Application Case:
Dawn S. Chandler et al successfully constructed a cell model simulating the genetic metabolic disorder phenylketonuria (PKU) in Hep G2 cells using CRISPR-Cas9 technology. The researchers precisely introduced predetermined point mutations via homology-directed repair (HDR) mechanisms to simulate the splicing defects of PKU. This method significantly enhanced the understanding of disease mechanisms and provided a valuable experimental model for the development of new treatment methods, laying a solid foundation for related studies and aiding in the development of effective intervention strategies for such diseases.
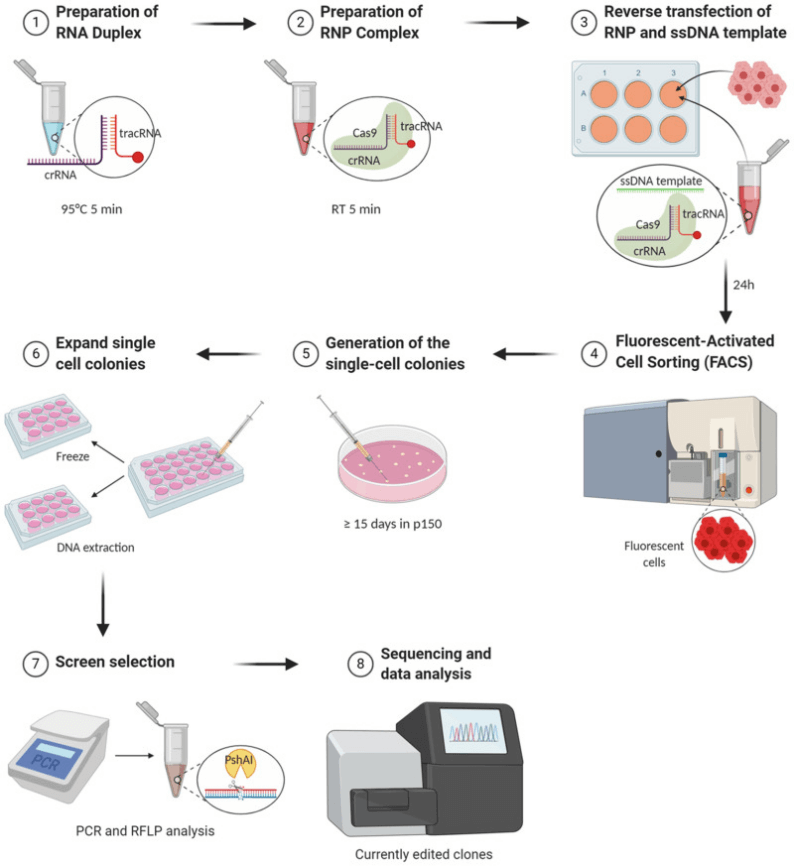
Figure 1. Outline of Gene Editing Experiment Plan
Catalog#: YC-D001
Source: The Huh7 cell line is derived from human hepatocellular carcinoma, initially established by Nakabayashi et al. in 1982 from a 57-year-old Japanese male liver tumor.
Molecular Expression: Huh7 cells express various genes related to liver function. Additionally, these cells can express genes associated with viral infection and immunity. For instance, certain host factor genes expressed in Huh7 cells are crucial for HCV infection and replication.
Growth Characteristics: Epithelial-like morphology, adherent growth. Displays resistance to various chemotherapeutic drugs.
Applications: Huh7 cells are a classic cell line for studying hepatitis C virus (HCV). They are also commonly used in research on the mechanisms of liver cancer development.
Application Case:
Ajay K Israni et al. employed CRISPR/Cas9 technology to precisely edit the CYP3A5 gene locus in Huh7 cells to study its gene expression and drug metabolism functions. First, the researchers selected Huh7 cells as hosts and designed specific guide RNAs (gRNA) targeting the 3 sites of CYP3A5. By successfully knocking out these sites, they obtained cell lines expressing CYP3A51. Drug metabolism analysis showed enhanced metabolic activity towards midazolam and tacrolimus in these cell lines, indicating that CYP3A5 significantly impacts drug metabolism. CYP3CIDE experiments further confirmed the different metabolic activities of these cell lines, providing new experimental models for studying the role of CYP3A5 in drug metabolism[2].
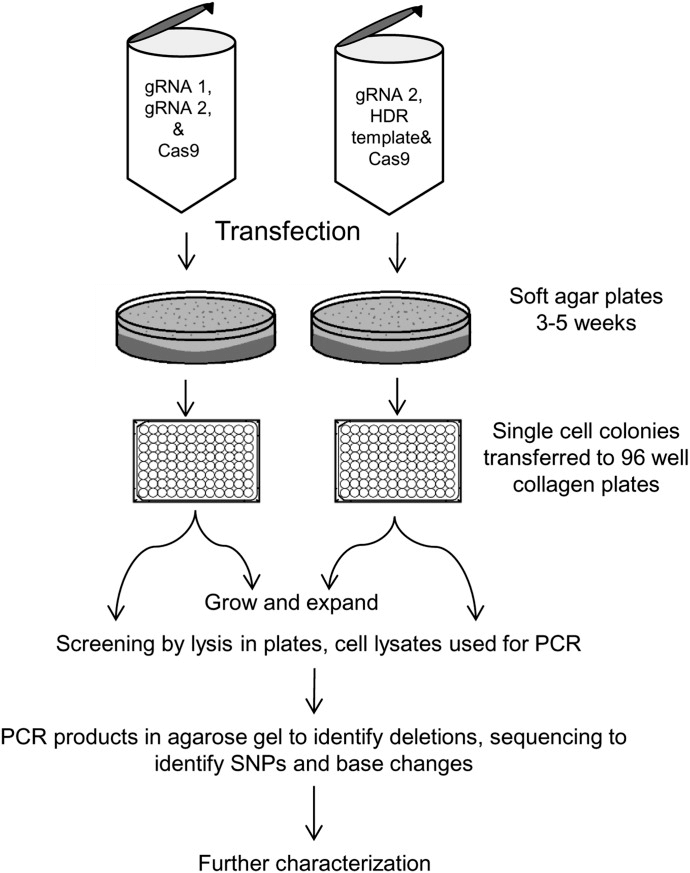
Figure 2. Workflow for Developing CYP3A5 Gene-modified Cell Lines Using CRISPR/Cas9 and Clonal Selection.
Catalog#: YC-C007
Source: Derived from human liver tissue.
Molecular Expression: SK-Hep1 cells express various tumor-related genes, such as the c-Myc gene. Additionally, SK-Hep1 cells significantly express the epidermal growth factor receptor (EGFR).
Growth Characteristics: Epithelial-like morphology, adherent growth. SK-Hep1 cells exhibit high metastatic potential and tumor formation ability.
Applications: SK-Hep1 cells are important models for researching the mechanisms of liver cancer occurrence and are also commonly used for screening and evaluating anti-tumor drugs.
Application Case:
Shuhua Zhang et al. utilized CRISPR-Cas9 gene editing technology to create NSD1 gene knockout SK-HEP-1 cell lines. The results showed that NSD1 was highly expressed in SK-HEP-1 cells and was related to poor prognosis in liver cancer patients. After knocking out NSD1, the proliferation, migration, and invasion abilities of SK-HEP-1 cells were inhibited, promoting H3K27me3 methylation and suppressing Wnt10b expression, which inhibited the Wnt/β-catenin signaling pathway. These changes were validated in a subcutaneous tumor model in nude mice, indicating that NSD1, H3, and Wnt10b may be potential therapeutic targets for hepatocellular carcinoma [3].
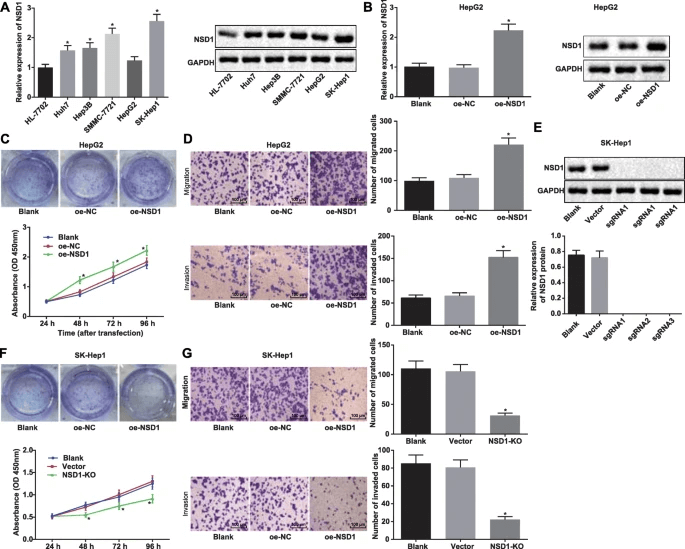
Figure 3. NSD1 knockout inhibits the proliferation, migration, and invasion capabilities of HCC cell lines.
In addition to these cell lines, there are other liver cancer cell models, such as Hep3B, SMMC-7721, and MHCC97H. You can choose the most appropriate cell model based on your research objectives, the genetic background of the cell line, biological characteristics, and drug metabolism properties to help the development of liver cancer treatment strategies and new drug screening.
Ubigene can provide gene editing (KO/KI/PM) and stable cell line customization services related to liver cancer research. Feel free to inquire!
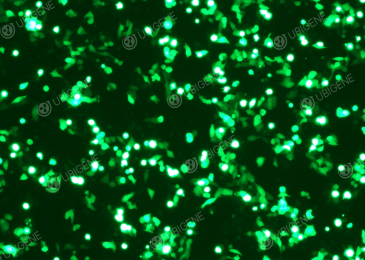
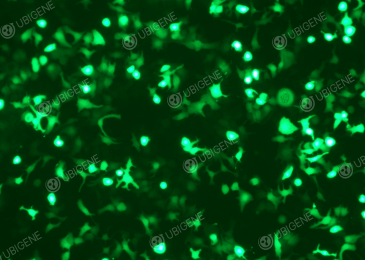
Selected Gene editing experiment figures
References
[1] Montes M, Sanford BL, Comiskey DF, Chandler DS. RNA splicing and disease: animal models to therapies. Trends Genet. 2019;35(1):68-87. https://doi.org/10.1016/j.tig.2018.10.002
[2] Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci U S A. 2011;108(27):11093-11098. https://doi.org/10.1073/pnas.1101135108
[3] Zhang S, Zhang F, Chen Q, Wan C, Xiong J, Xu J. CRISPR/Cas9-mediated knockout of NSD1 suppresses the hepatocellular carcinoma development via the NSD1/H3/Wnt10b signaling pathway. J Exp Clin Cancer Res. 2019;38:467. https://doi.org/10.1186/s13046-019-1462-y






Expert Insights | Cell Models Commonly Used in Liver Cancer Research

Liver cancer, or primary liver cancer, is a prevalent malignant tumor worldwide. This disease typically shows no obvious symptoms in the early stages and progresses rapidly, with most patients being diagnosed at an advanced stage. Symptoms of liver cancer may include pain in the liver area, fatigue, weight loss, jaundice, and ascites. Cell models of liver cancer are crucial for studying the occurrence, development, and biological characteristics of the disease, providing essential experimental platforms for drug screening, treatment evaluation, and new drug development. Today, we will share some commonly used cell models in liver cancer research and their application cases.
Catalog#: YC-C001
Source: The Hep G2 cell line is derived from a 15-year-old Caucasian male with hepatoblastoma.
Molecular Expression: Hep G2 cells retain many genetic expression features of normal liver cells and can express various liver-specific genes.
Growth Characteristics: Similar morphology to normal liver cells, adherent growth.
Applications: Widely used in various liver-related research areas, including drug toxicity assessments, bioartificial liver system construction, and studies on hepatitis B virus models.
Application Case:
Dawn S. Chandler et al successfully constructed a cell model simulating the genetic metabolic disorder phenylketonuria (PKU) in Hep G2 cells using CRISPR-Cas9 technology. The researchers precisely introduced predetermined point mutations via homology-directed repair (HDR) mechanisms to simulate the splicing defects of PKU. This method significantly enhanced the understanding of disease mechanisms and provided a valuable experimental model for the development of new treatment methods, laying a solid foundation for related studies and aiding in the development of effective intervention strategies for such diseases.

Figure 1. Outline of Gene Editing Experiment Plan
Catalog#: YC-D001
Source: The Huh7 cell line is derived from human hepatocellular carcinoma, initially established by Nakabayashi et al. in 1982 from a 57-year-old Japanese male liver tumor.
Molecular Expression: Huh7 cells express various genes related to liver function. Additionally, these cells can express genes associated with viral infection and immunity. For instance, certain host factor genes expressed in Huh7 cells are crucial for HCV infection and replication.
Growth Characteristics: Epithelial-like morphology, adherent growth. Displays resistance to various chemotherapeutic drugs.
Applications: Huh7 cells are a classic cell line for studying hepatitis C virus (HCV). They are also commonly used in research on the mechanisms of liver cancer development.
Application Case:
Ajay K Israni et al. employed CRISPR/Cas9 technology to precisely edit the CYP3A5 gene locus in Huh7 cells to study its gene expression and drug metabolism functions. First, the researchers selected Huh7 cells as hosts and designed specific guide RNAs (gRNA) targeting the 3 sites of CYP3A5. By successfully knocking out these sites, they obtained cell lines expressing CYP3A51. Drug metabolism analysis showed enhanced metabolic activity towards midazolam and tacrolimus in these cell lines, indicating that CYP3A5 significantly impacts drug metabolism. CYP3CIDE experiments further confirmed the different metabolic activities of these cell lines, providing new experimental models for studying the role of CYP3A5 in drug metabolism[2].

Figure 2. Workflow for Developing CYP3A5 Gene-modified Cell Lines Using CRISPR/Cas9 and Clonal Selection.
Catalog#: YC-C007
Source: Derived from human liver tissue.
Molecular Expression: SK-Hep1 cells express various tumor-related genes, such as the c-Myc gene. Additionally, SK-Hep1 cells significantly express the epidermal growth factor receptor (EGFR).
Growth Characteristics: Epithelial-like morphology, adherent growth. SK-Hep1 cells exhibit high metastatic potential and tumor formation ability.
Applications: SK-Hep1 cells are important models for researching the mechanisms of liver cancer occurrence and are also commonly used for screening and evaluating anti-tumor drugs.
Application Case:
Shuhua Zhang et al. utilized CRISPR-Cas9 gene editing technology to create NSD1 gene knockout SK-HEP-1 cell lines. The results showed that NSD1 was highly expressed in SK-HEP-1 cells and was related to poor prognosis in liver cancer patients. After knocking out NSD1, the proliferation, migration, and invasion abilities of SK-HEP-1 cells were inhibited, promoting H3K27me3 methylation and suppressing Wnt10b expression, which inhibited the Wnt/β-catenin signaling pathway. These changes were validated in a subcutaneous tumor model in nude mice, indicating that NSD1, H3, and Wnt10b may be potential therapeutic targets for hepatocellular carcinoma [3].

Figure 3. NSD1 knockout inhibits the proliferation, migration, and invasion capabilities of HCC cell lines.
In addition to these cell lines, there are other liver cancer cell models, such as Hep3B, SMMC-7721, and MHCC97H. You can choose the most appropriate cell model based on your research objectives, the genetic background of the cell line, biological characteristics, and drug metabolism properties to help the development of liver cancer treatment strategies and new drug screening.
Ubigene can provide gene editing (KO/KI/PM) and stable cell line customization services related to liver cancer research. Feel free to inquire!


Selected Gene editing experiment figures
References
[1] Montes M, Sanford BL, Comiskey DF, Chandler DS. RNA splicing and disease: animal models to therapies. Trends Genet. 2019;35(1):68-87. https://doi.org/10.1016/j.tig.2018.10.002
[2] Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci U S A. 2011;108(27):11093-11098. https://doi.org/10.1073/pnas.1101135108
[3] Zhang S, Zhang F, Chen Q, Wan C, Xiong J, Xu J. CRISPR/Cas9-mediated knockout of NSD1 suppresses the hepatocellular carcinoma development via the NSD1/H3/Wnt10b signaling pathway. J Exp Clin Cancer Res. 2019;38:467. https://doi.org/10.1186/s13046-019-1462-y





