Location:Home > Application > CRISPR Library: Opening a New Perspective for Virus Research - Special Guide (Part 1)
CRISPR Library: Opening a New Perspective for Virus Research - Special Guide (Part 1)
On the scientific research path to combat viruses, CRISPR libraries, with their powerful functionality, reveal the interactions between viruses and host cells. Using the application of CRISPR libraries in influenza virus research as an example, the following content will explore how CRISPR libraries open new perspectives for virus research.
Influenza virus poses a significant threat to global public health, leading to a large number of illnesses and deaths annually. Its rapid mutational characteristic challenges vaccine development. Hence, there is a need to find more effective and broad-spectrum anti-influenza strategies. In recent years, host-directed antiviral therapy has garnered attention. This approach does not directly target the virus itself but instead adjusts the characteristics of host cells to restrict viral infection and replication, demonstrating potential broad-spectrum and antiviral resistance advantages, providing a new direction for anti-influenza strategies.
Trimarco et al. employed CRISPR activation library screening technology to identify host factors that can restrict influenza virus. They used influenza B virus (IBV) to treat cells and then used the CRISPR-Cas9 system to activate host genes, searching for genes that could enhance the cell's resistance to viral infection. Through this method, they discovered the B3GAT1 gene, which encodes an enzyme called β-1,3-glucuronyltransferase 1, capable of altering the cell's surface glycan structure, thus affecting viral infection.
Library Type: Human genome-wide activation CRISPR library (Addgene 92379)
Transduced Cells: A549 cells
Virus: B/Yamagata/16/1988 (Yam/88) strain of IBV
Screening Strategy: Three rounds of virus infection, positive selection
Screening Method:
Each replicate collected half of the stable transduced cells from the library and stored them at -80°C. The remaining half of the cells in each replicate were infected with the B/Yamagata/16/1988 and then incubated for 24 hours in medium containing TPCK-treated trypsin to promote multi-cycle infection. Subsequently, the medium was replaced with DMEM medium, and the cells were cultured for 48 hours to fully exhibit the cellular cytopathic effects from viral infection. The cells were then trypsinized, replated to remove dead cells, and the culture continued. When the cells reached the initial cell amount, half of the cells were collected, lysed, and stored, and the remaining half of the cells were infected with the virus again. This cell culture, cell collection, and re-infection process were repeated two more times for a total of three rounds of infection until both replicates of cells showed no obvious cellular cytopathic effects after infection. The genomic DNA of the cells after each round of infection was extracted, and deep sequencing was performed on the sgRNAs using high-throughput sequencing technology. The enriched sgRNAs were identified through bioinformatics analysis of the sequencing data, thereby determining candidate antiviral host genes.
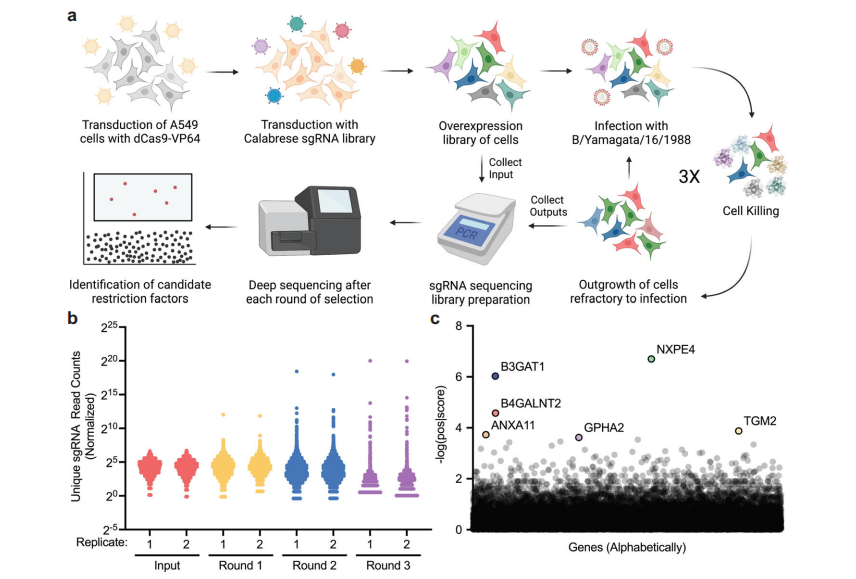
Figure 1: Determination of the Restrictive Factors of Influenza B Virus through Genome-wide Activation CRISPR Library Screening
The researchers hypothesized that repeated infections would lead to internal competition among enriched genes, eventually resulting in a bottleneck effect on the population, containing only a few genes with strong antiviral activity. As expected, the number of detected sgRNAs decreased in each round of infection, and the count of a few sgRNAs became increasingly prominent in the population.
For the most promising screening results, the researchers selected the top six enriched sgRNAs targeting genes for validation. Among them, the B3GAT1 gene, previously not explicitly described as a viral restriction factor, exhibited significant antiviral activity. Therefore, the researchers chose the B3GAT1 gene for in-depth characterization studies.
The researchers observed inhibited viral infection in B3GAT1 overexpressing A549 cells using flow cytometry and immunofluorescence microscopy.
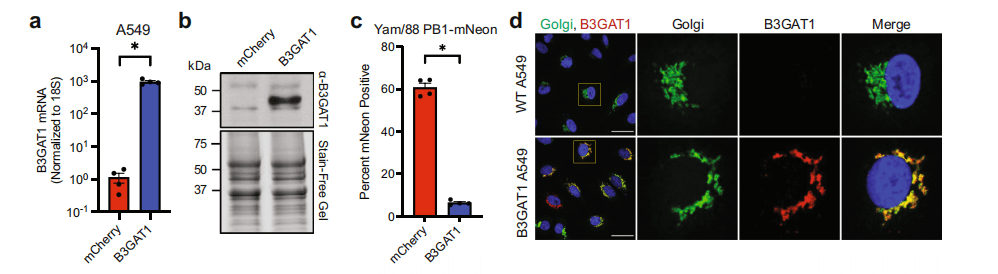
Figure 2: Validation of B3GAT1 overexpressing A549 cells
B3GAT1 is a glycosyltransferase. The researchers used MALDI-TOF mass spectrometry to analyze the N-linked glycans of B3GAT1-overexpressing cells and control cells. They found a reduction in the types of glycans containing sialic acid and an increase in the types of glycans containing glucuronic acid (GlcA) in B3GAT1-overexpressing cells. Using different fluorescently labeled lectins and specific antibodies to detect the expression of sialic acid and GlcA on the cell surface, they found reduced expression of α2,3- and α2,6-linked sialic acid in B3GAT1-expressing cells. Notably, many viruses require sialic acid for host cell entry.
Based on these findings, the researchers proposed a hypothesis: B3GAT1 competes with host cell sialyltransferases, preventing the surface expression of sialic acid on the cell, thus impeding viral receptor binding and subsequent cell entry.
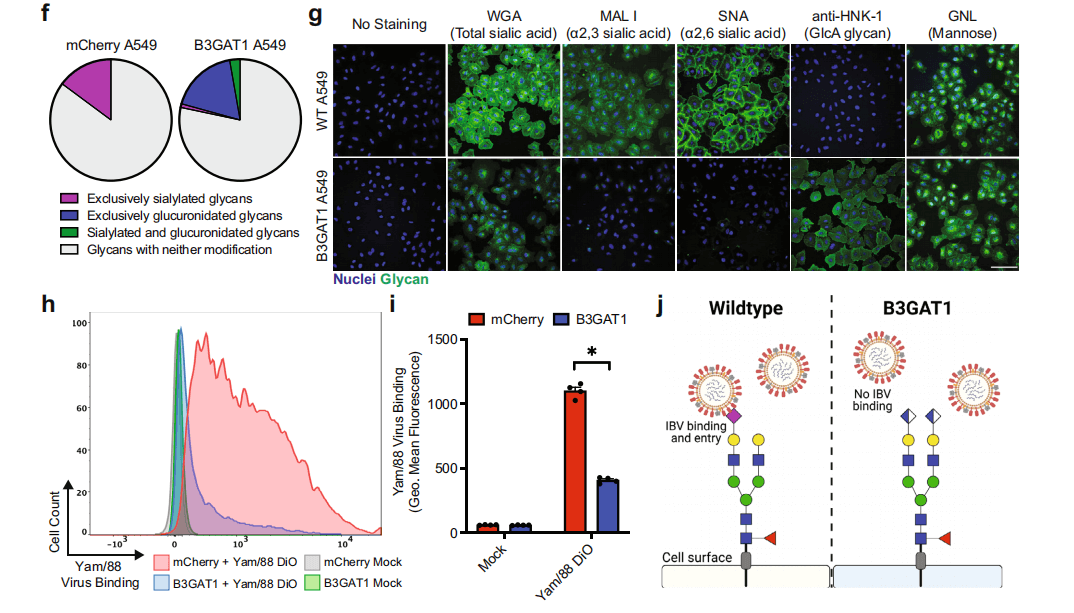
Figure 3: B3GAT1 can prevent the surface expression of sialic acid by competing with sialyltransferases.
The researchers further tested the antiviral effect of B3GAT1 on various influenza viruses, including the Victoria and Yamagata lineages of influenza B virus, as well as the H1N1 and H3N2 subtypes of influenza A virus. In a mouse model, through adeno-associated virus (AAV) vector-mediated specific expression of B3GAT1 in mouse respiratory epithelia, they evaluated its protective effect against challenge with influenza viruses. The results showed that B3GAT1, by altering the cell's surface glycan composition, particularly by reducing sialic acid expression, broadly restricted viruses that rely on sialic acid for host cell entry.
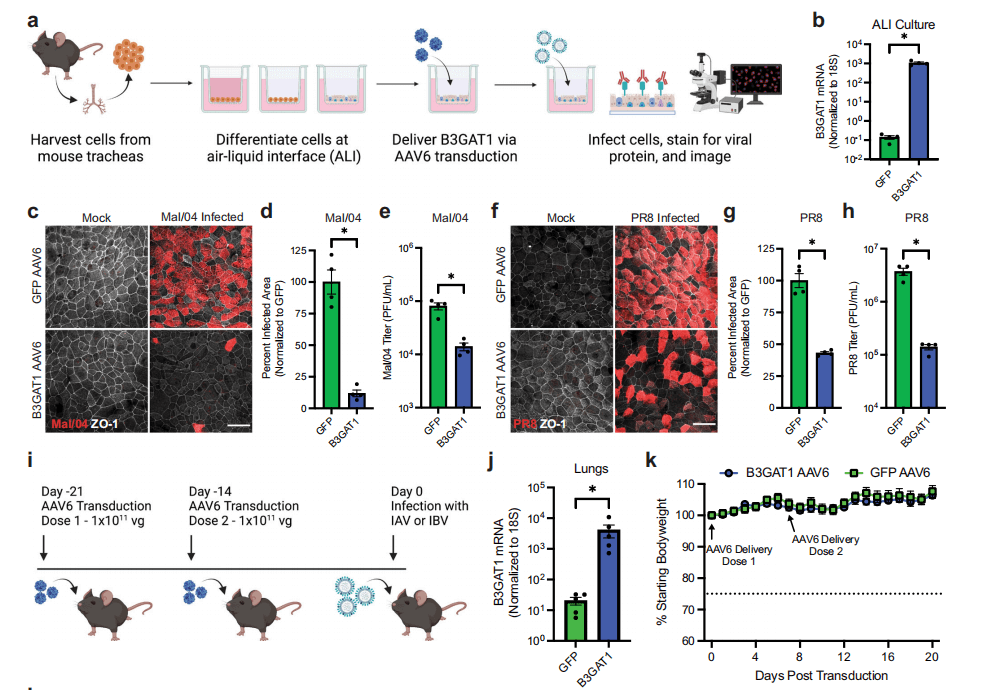
Figure 4: Safe in vivo expression of B3GAT1 provides protection against lethal influenza virus infection.
This study found that B3GAT1 effectively restricts the infection of various influenza viruses and other viruses by altering the surface glycan structure of host cells. The study also demonstrated that B3GAT1 overexpression in mice not only is well-tolerated but also provides protection against lethal influenza virus challenges, offering a new direction for the development of novel broad-spectrum antiviral therapies.
The CRISPR library screening technology has been applied to a variety of virus research, including influenza virus, HIV, Flaviviruses (such as dengue virus, Zika virus, West Nile virus), coronaviruses (such as SARS-CoV-2), hepatitis B virus, hepatitis A virus, hepatitis C virus, herpes virus, etc. These CRISPR library screenings have revealed a variety of factors critical to the virus lifecycle within host cells, including virus entry, replication, assembly, and host immune response, providing important molecular targets and in-depth mechanistic understanding for antiviral drug development and vaccine design.
Ubigene can provide in-stock plasmids and virus particles for over 35 libraries, including the human genome-wide activation library used in the study. The library virus particles are available for as low as $1845, and there are more types and deliverables available at promotional prices. Feel free to contact us for more details.
Reference
[1] Trimarco, Joseph D et al. “Cellular glycan modification by B3GAT1 broadly restricts influenza virus infection.” Nature communications vol. 13,1 6456. 29 Oct. 2022, doi:10.1038/s41467-022-34111-0
[2] [2]Srivastava, Kaveri, and Bhaswati Pandit. “Genome-wide CRISPR screens and their applications in infectious disease.” Frontiers in genome editing vol. 5 1243731. 19 Sep. 2023, doi:10.3389/fgeed.2023.1243731
[3]Chulanov, Vladimir et al. “CRISPR Screening: Molecular Tools for Studying Virus-Host Interactions.” Viruses vol. 13,11 2258. 11 Nov. 2021, doi:10.3390/v13112258


CRISPR Library: Opening a New Perspective for Virus Research - Special Guide (Part 1)

On the scientific research path to combat viruses, CRISPR libraries, with their powerful functionality, reveal the interactions between viruses and host cells. Using the application of CRISPR libraries in influenza virus research as an example, the following content will explore how CRISPR libraries open new perspectives for virus research.
Influenza virus poses a significant threat to global public health, leading to a large number of illnesses and deaths annually. Its rapid mutational characteristic challenges vaccine development. Hence, there is a need to find more effective and broad-spectrum anti-influenza strategies. In recent years, host-directed antiviral therapy has garnered attention. This approach does not directly target the virus itself but instead adjusts the characteristics of host cells to restrict viral infection and replication, demonstrating potential broad-spectrum and antiviral resistance advantages, providing a new direction for anti-influenza strategies.
Trimarco et al. employed CRISPR activation library screening technology to identify host factors that can restrict influenza virus. They used influenza B virus (IBV) to treat cells and then used the CRISPR-Cas9 system to activate host genes, searching for genes that could enhance the cell's resistance to viral infection. Through this method, they discovered the B3GAT1 gene, which encodes an enzyme called β-1,3-glucuronyltransferase 1, capable of altering the cell's surface glycan structure, thus affecting viral infection.
Library Type: Human genome-wide activation CRISPR library (Addgene 92379)
Transduced Cells: A549 cells
Virus: B/Yamagata/16/1988 (Yam/88) strain of IBV
Screening Strategy: Three rounds of virus infection, positive selection
Screening Method:
Each replicate collected half of the stable transduced cells from the library and stored them at -80°C. The remaining half of the cells in each replicate were infected with the B/Yamagata/16/1988 and then incubated for 24 hours in medium containing TPCK-treated trypsin to promote multi-cycle infection. Subsequently, the medium was replaced with DMEM medium, and the cells were cultured for 48 hours to fully exhibit the cellular cytopathic effects from viral infection. The cells were then trypsinized, replated to remove dead cells, and the culture continued. When the cells reached the initial cell amount, half of the cells were collected, lysed, and stored, and the remaining half of the cells were infected with the virus again. This cell culture, cell collection, and re-infection process were repeated two more times for a total of three rounds of infection until both replicates of cells showed no obvious cellular cytopathic effects after infection. The genomic DNA of the cells after each round of infection was extracted, and deep sequencing was performed on the sgRNAs using high-throughput sequencing technology. The enriched sgRNAs were identified through bioinformatics analysis of the sequencing data, thereby determining candidate antiviral host genes.

Figure 1: Determination of the Restrictive Factors of Influenza B Virus through Genome-wide Activation CRISPR Library Screening
The researchers hypothesized that repeated infections would lead to internal competition among enriched genes, eventually resulting in a bottleneck effect on the population, containing only a few genes with strong antiviral activity. As expected, the number of detected sgRNAs decreased in each round of infection, and the count of a few sgRNAs became increasingly prominent in the population.
For the most promising screening results, the researchers selected the top six enriched sgRNAs targeting genes for validation. Among them, the B3GAT1 gene, previously not explicitly described as a viral restriction factor, exhibited significant antiviral activity. Therefore, the researchers chose the B3GAT1 gene for in-depth characterization studies.
The researchers observed inhibited viral infection in B3GAT1 overexpressing A549 cells using flow cytometry and immunofluorescence microscopy.

Figure 2: Validation of B3GAT1 overexpressing A549 cells
B3GAT1 is a glycosyltransferase. The researchers used MALDI-TOF mass spectrometry to analyze the N-linked glycans of B3GAT1-overexpressing cells and control cells. They found a reduction in the types of glycans containing sialic acid and an increase in the types of glycans containing glucuronic acid (GlcA) in B3GAT1-overexpressing cells. Using different fluorescently labeled lectins and specific antibodies to detect the expression of sialic acid and GlcA on the cell surface, they found reduced expression of α2,3- and α2,6-linked sialic acid in B3GAT1-expressing cells. Notably, many viruses require sialic acid for host cell entry.
Based on these findings, the researchers proposed a hypothesis: B3GAT1 competes with host cell sialyltransferases, preventing the surface expression of sialic acid on the cell, thus impeding viral receptor binding and subsequent cell entry.

Figure 3: B3GAT1 can prevent the surface expression of sialic acid by competing with sialyltransferases.
The researchers further tested the antiviral effect of B3GAT1 on various influenza viruses, including the Victoria and Yamagata lineages of influenza B virus, as well as the H1N1 and H3N2 subtypes of influenza A virus. In a mouse model, through adeno-associated virus (AAV) vector-mediated specific expression of B3GAT1 in mouse respiratory epithelia, they evaluated its protective effect against challenge with influenza viruses. The results showed that B3GAT1, by altering the cell's surface glycan composition, particularly by reducing sialic acid expression, broadly restricted viruses that rely on sialic acid for host cell entry.

Figure 4: Safe in vivo expression of B3GAT1 provides protection against lethal influenza virus infection.
This study found that B3GAT1 effectively restricts the infection of various influenza viruses and other viruses by altering the surface glycan structure of host cells. The study also demonstrated that B3GAT1 overexpression in mice not only is well-tolerated but also provides protection against lethal influenza virus challenges, offering a new direction for the development of novel broad-spectrum antiviral therapies.
The CRISPR library screening technology has been applied to a variety of virus research, including influenza virus, HIV, Flaviviruses (such as dengue virus, Zika virus, West Nile virus), coronaviruses (such as SARS-CoV-2), hepatitis B virus, hepatitis A virus, hepatitis C virus, herpes virus, etc. These CRISPR library screenings have revealed a variety of factors critical to the virus lifecycle within host cells, including virus entry, replication, assembly, and host immune response, providing important molecular targets and in-depth mechanistic understanding for antiviral drug development and vaccine design.
Ubigene can provide in-stock plasmids and virus particles for over 35 libraries, including the human genome-wide activation library used in the study. The library virus particles are available for as low as $1845, and there are more types and deliverables available at promotional prices. Feel free to contact us for more details.
Reference
[1] Trimarco, Joseph D et al. “Cellular glycan modification by B3GAT1 broadly restricts influenza virus infection.” Nature communications vol. 13,1 6456. 29 Oct. 2022, doi:10.1038/s41467-022-34111-0
[2] [2]Srivastava, Kaveri, and Bhaswati Pandit. “Genome-wide CRISPR screens and their applications in infectious disease.” Frontiers in genome editing vol. 5 1243731. 19 Sep. 2023, doi:10.3389/fgeed.2023.1243731
[3]Chulanov, Vladimir et al. “CRISPR Screening: Molecular Tools for Studying Virus-Host Interactions.” Viruses vol. 13,11 2258. 11 Nov. 2021, doi:10.3390/v13112258


