Luciferase Assay: Principles, Purpose, and Process

Luciferase Assay: Principles, Purpose, and Process

Contents
What is the principle of luciferase assay?
Luciferases are a class of enzymes that generate bioluminescent signals by oxidizing luciferin substrates. Examples include Firefly Luciferase (FLuc), Renilla Luciferase (RLuc), and Gaussia Luciferase (GLuc). Luciferase reporter assays are widely applied in various fields of biological research, such as gene expression and regulation studies, tumor cell tracking, etc. FLuc catalyzes the reaction of its substrate luciferin in the presence of ATP and Mg2+, producing yellow-green light with a wavelength of 550-570 nm. In contrast, RLuc only requires oxygen to catalyze the substrate coelenterazine, resulting in blue light at approximately 480 nm. GLuc also uses coelenterazine as its substrate, and GLuc is a secretory protein, allowing the collection of cell supernatants for subsequent luminescence detection.
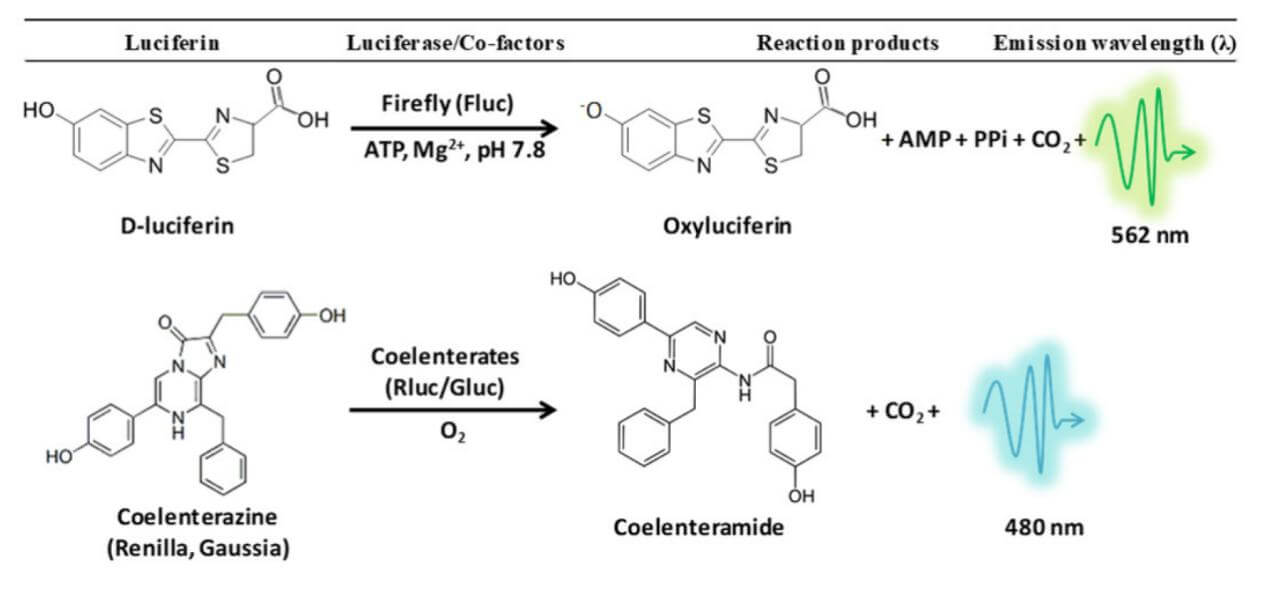
Figure 1. Experimental Principle of Luciferase Reporter Assay
Luciferase reporter assays generally refer to systems that detect the regulatory capabilities of gene regulatory elements through luciferase activity, including single luciferase reporter assay system and dual luciferase reporter assay system. The single luciferase reporter system is easy to operate and is commonly used to study the functional effects of gene regulatory elements in stably transfected cells. However, it cannot differentiate whether differences in results between experimental groups arise from specific regulation of the reporter gene or global cell events (such as proliferation, apoptosis, necrosis, etc.). Before analyzing experimental results, it is essential to normalize the data. In transient transfection experiments, significant variances may be introduced due to transfection efficiency, which cannot be normalized through cell number alone; therefore, a reference gene is necessary to assess transfection efficiency, which makes the experimental data more accurate. This system is known as the dual luciferase reporter system. FLuc and RLuc are commonly used in the dual luciferase reporter system due to their short half-lives, distinct substrates, different luminescent colors, and RLuc's relative insensitivity to cellular environment impacts, making it suitable as a reference gene.
What are the advantages of luciferase assays?
Different reporter systems have unique characteristics that are suitable for various applications. In Chloramphenicol Acetyltransferase (CAT) assays, CAT is not expressed in eukaryotic cells, and the protein is highly stable, offering high detection sensitivity. However, the CAT assay can be cumbersome to perform and has a narrow linear range. E. coliβ-galactosidase is often used as a reference gene for other reporter factors, but its activity is relatively high in bacteria and serum. In the case of fluorescent protein reporter systems, proteins can be directly observed under excitation light, allowing for long-term monitoring of live cell protein localization and quantification. Still, many substances in organisms produce non-specific fluorescence when exposed to excitation light (such as mouse fur and skin), affecting detection sensitivity. Conversely, bioluminescent reporter assays rely on the specificity of the enzyme-substrate reaction for luminescence, eliminating the need for excitation light, and luciferases are typically not expressed in most cells, resulting in extremely low background. Additionally, firefly luciferase emits light with strong penetration, making it suitable for detecting the localization and quantification of transplanted cells within an organism.
| Reporter Gene | Application Scenario |
| Chloramphenicol acetyltransferase (CAT) | In vitro experiments: Gene expression. |
| E. coli β-galactosidase | In vitro experiments: Reference gene, immunohistochemistry. |
| Luciferase | Dual luciferase reporter system; in vivo experiments. |
| Fluorescent protein | Live cell observation. |
Table 1. Comparison Analysis of Different Reporter Systems
What is the purpose of the luciferase assay?
Luciferase reporter assays have widespread potential applications including the study of interactions between transcription factors and target gene promoters, interactions between microRNA and mRNA, detection of signaling pathway activities, and high-throughput screening of chemical compounds on protein expression.

Figure 2. Schematic Diagram of Studying the Interaction between Transcription Factors and Target Gene Promoters
Transcription Factor and Target Gene Promoter Interactions
Clone the target gene promoter region (~2000 bp) into the upstream region of the FLuc gene and co-transfect with the corresponding transcription factor to detect luciferase activity.
MicroRNA and mRNA Interactions
Clone the 3' UTR of the target gene into the 3' UTR region of the FLuc gene and co-transfect the corresponding regulatory microRNA to assess luciferase activity.
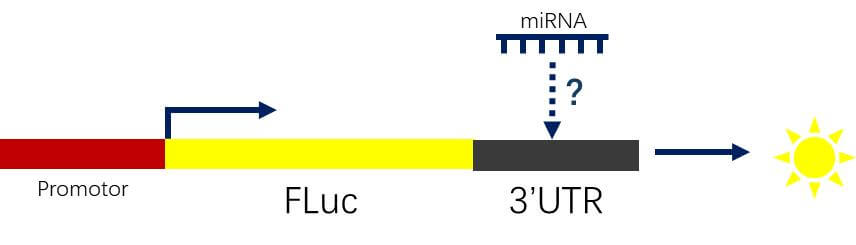
Figure 3. Schematic Diagram of miRNA mRNA Interaction
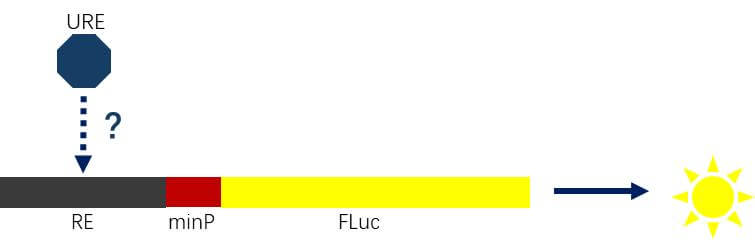
Figure 4. Schematic Diagram of the Activity of the Detection Signal Pathway
Detection of Signaling Pathway Activity
Clone the response element (RE) sequence into a minimal promoter (minP) upstream and co-transfect with the corresponding upstream regulatory element (URE) to study whether the signaling pathway is activated.
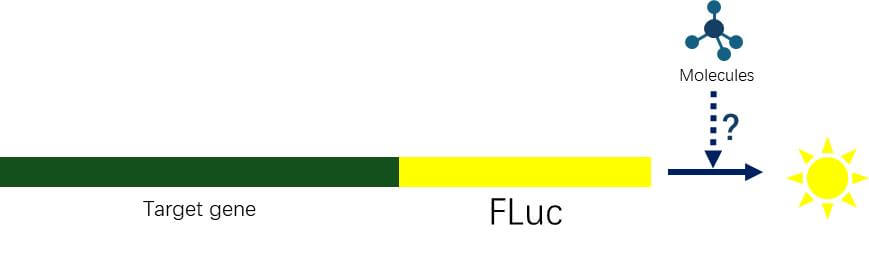
Figure 5. Schematic Diagram of the Effect of High-throughput Screening of Small Molecules on Protein Expression
High-Throughput Screening of Small Molecules on Protein Expression
Using CRISPR-CAS9 technology, knock the luciferase gene into the 3' end of the target gene, forming a fusion protein with the target protein. Assess the impact of specific small molecules on target protein expression through luciferase activity, suitable for low-expression protein studies.
How do you perform a luciferase assay?
Step 1: Plasmid Construction
Choose a suitable plasmid based on the research objectives. When studying the effects of transcription factors on gene expression, pGL4.20 and pRL vectors are typically used. The pGL4.20 vector has the FLuc gene upstream of a multiple cloning site without a promoter, allowing the insertion of a promoter sequence of interest. The pRL series of vectors has different promoters; an appropriate promoter should be chosen based on experimental needs, typically selecting a weakly active TK promoter.
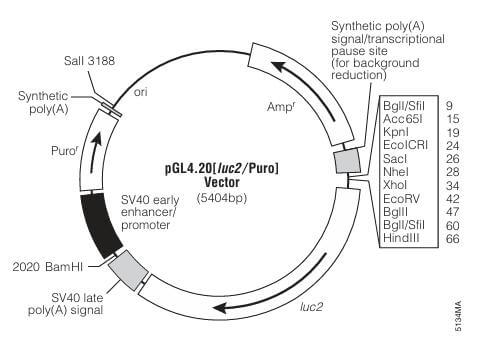
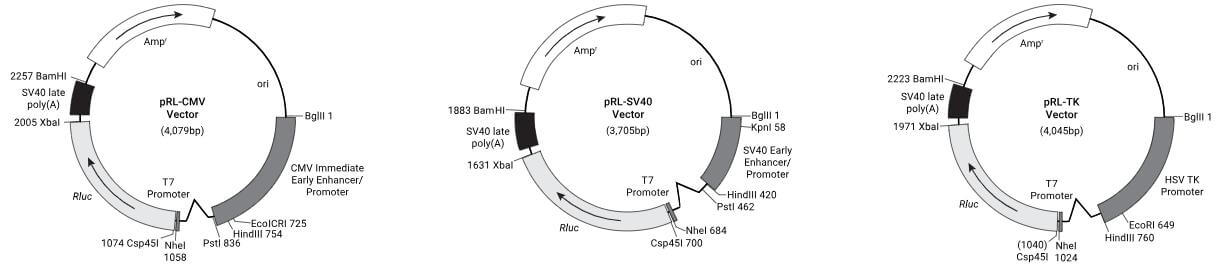
Figure 6. Schematic Diagram of pGL4.20 Vector and pRL Series Vector
In studying microRNA and mRNA interactions, pMIR-REPORT Luciferase can be selected, which contains a multiple cloning site at the 3' UTR of FLuc for inserting the target gene's 3' UTR. The reference for this plasmid is the pMIR-REPORT™ β-galactosidase reporter gene control vector. Additionally, the reference gene and the reporter gene may be placed on the same plasmid, named pmirGLO, for assessing miRNA activity.
Step 2: Cell Transfection
Transfection reagents may include PEI, calcium chloride, or lipofectamine 2000. If using the dual plasmid system, the ratio of reference gene plasmid to reporter gene plasmid should be approximately 1:10 to ensure that the reference gene does not interfere with the expression of the reporter gene. Detection should occur 24-36 hours post-transfection.
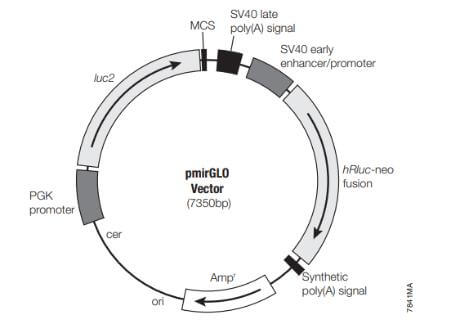
Figure 7. Schematic Diagram of pmirGLOP Vector
Step 3: Reporter Gene Detection
Lyse the cells and centrifuge to collect the culture medium supernatant, placing it in a 96-well plate. Add the appropriate FLuc buffer and measure luminescence. Then add STOP+RLuc buffer and measure luminescence again.
Luciferase Assay Data Analysis
1. Calculate the ratio of Firefly Luciferase/Renilla Luciferase luminescent intensity for each well.
2. Compute the average value (Average-control) of multiple replicates from the control group, and calculate the normalized value by dividing the fluorescence ratio of the experimental group by this average, treating the control group's value as 1 for data analysis.
3. Analyze the normalized values of the control and experimental groups by creating a bar graph.
Luciferase Assay Troubleshooting
1. High Luminescence Values
Excessive luminescence may exceed the detection range of the instrument; consider reducing the quantity of transfected plasmids and cell numbers, or diluting the lysate.
2. Low Luminescence Values
Luminescence close to background levels may cause significant result errors; consider increasing the amount of transfected plasmids and cell amount or decreasing the volume of the lysate. Because luciferase reporter assays are enzyme-catalyzed reactions, it is vital to ensure that the substrate quantity is in excess and to balance all components of the reaction to room temperature (optimal reaction temperature).
3. Large Variation Between Replicates
If using a multichannel pipette, ensure each tip is firmly inserted, and avoid bubble formation during the operation. For lysate samples, centrifuge before taking the supernatant to ensure sample homogeneity.
For more luciferase cells in-stock products, feel free to inquire!


