Target Screening: CRISPR Library vs Traditional Omics

Target Screening: CRISPR Library vs Traditional Omics

Target screening is a crucial task in the fields of life science research and precision medicine. It provides key directions for the diagnosis, treatment, and drug development of diseases. The emergence of CRISPR technology undoubtedly brought revolutionary changes to this field. There are various methods for target screening, including traditional omics methodologies and CRISPR library screening methodologies, but each method has its own advantages and limitations.
Contents
Traditional omics methodologies for targetscreening
Traditional omics methodologies play an important role in drug target screening, mainly including genomics, transcriptomics, proteomics, and metabolomics. These methodologies reveal the changes in biological systems under different physiological or pathological states through large-scale detection and analysis of genes, transcripts, proteins, or metabolites in biological samples, to identify potential disease-related targets.
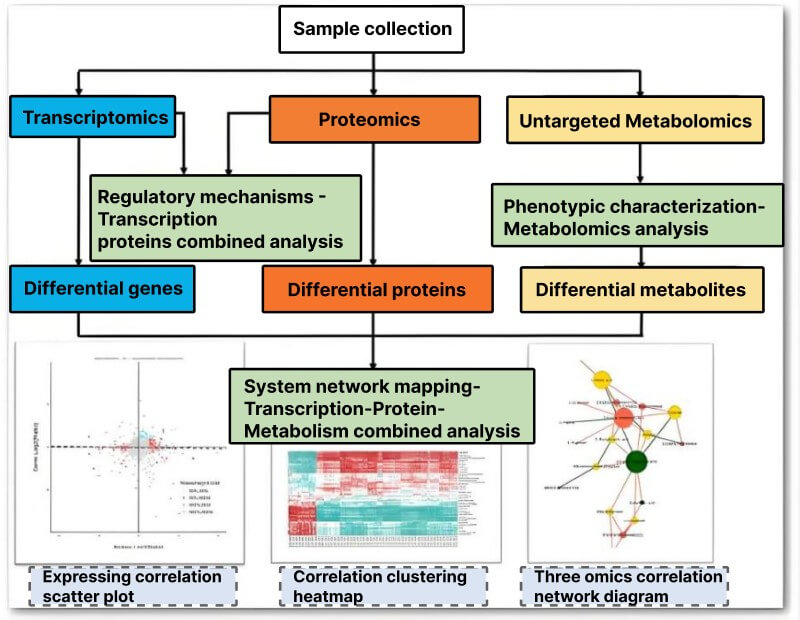
Figure 1. Traditional omics methodologies for target screening
Genomics:
Focuses on studying the entire genomic information of an organism. For example, genome-wide association studies (GWAS) compares the genome sequences of a large number of cases and control populations to identify single nucleotide polymorphisms (SNP) that are significantly associated with disease traits. These associated sites may reside in coding regions or regulatory regions of genes, thereby affecting the function or expression level of genes and being associated with the occurrence and development of diseases.
Transcriptomics:
Primarily studies the collection of all RNA transcripts transcribed by cells under specific conditions. Through RNA sequencing (RNA-seq) technology, gene expression level information can be obtained to analyze differentially expressed genes between disease tissues and normal tissues.
Proteomics:
The study of protein composition, expression levels, modification states, and interactions between proteins in cells or tissues. Proteomic analysis based on mass spectrometry technology can identify a large number of proteins and quantitatively analyze their differences in different samples.
Metabolomics:
The study of the types, quantities, and variations of metabolites in organisms. By detecting metabolites in samples such as blood, urine, and tissues using techniques such as gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS), it is possible to detect metabolic pathway disorders in disease states.
CRISPR Library Screening
CRISPR/Cas9 library screening technology is an efficient method for studying gene function. By constructing a large library of guide RNAs (gRNAs) targeting different genes and introducing them into cell populations, the genome is edited using Cas9 protein. Then, through specific screening strategies such as cell survival, proliferation, phenotype changes, etc., genes related to specific biological processes or diseases can be identified.
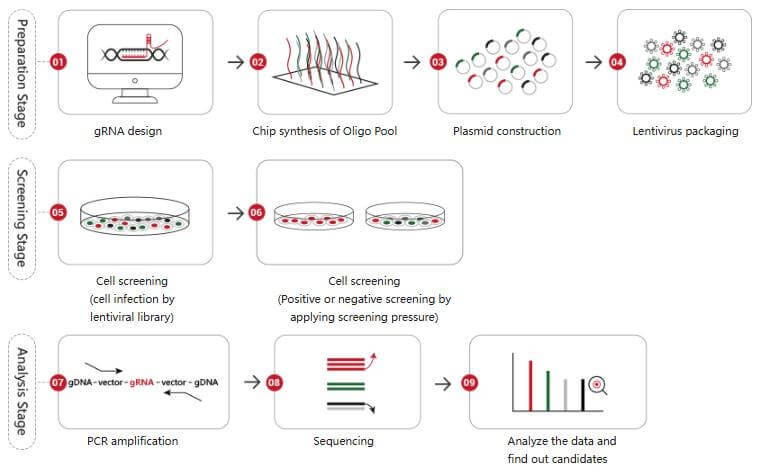
Figure 2. CRISPR screening workflow
CRISPR Screen vs Traditional Omics
| CRISPR Screen | Traditional Omics | |
| Advantage | 1. Directly evaluate gene function 2. High-throughput functional verification 3. Able to discover new drug targets 4. Quickly verify causal relationships | 1. Can obtain a systematic molecular map 2. Suitable for clinical sample analysis 3. Able to discover new molecular subtypes 4. Help to understand disease heterogeneity |
| Limitation | 1. Mainly limited to in vitro experiments 2. Off-target effects may occur 3. High requirements for library design and quality control 4. Some genes may be difficult to edit | 1. Correlation analysis is difficult to determine causal relationships, and data analysis is complex 2. Requires strong bioinformatics support 3. Long functional verification cycle |
Advantages of CRISPR Library Screening
1. In terms of functional verification
The CRISPR library screening technology can directly study the functional loss or acquisition of genes, and clarify the causal relationship of genes in specific biological processes.
2. Comprehensive screening
CRISPR library screening can perform unbiased screening across the entire genome, theoretically identifying any genes related to screening conditions, including some genes that have not been studied or whose functions are unknown.
3. Target operability
Compared to traditional omics methods that screen for targets such as proteins and metabolites, which require complex drug design and development processes (such as developing protein target small molecule inhibitors or antibody drugs), CRISPR library screening identifies targets that are mostly directly editable genes, making it easier for subsequent gene therapy drug development. For example, for single gene inherited disease pathogenic genes, gene editing strategies can be directly developed.
Application: You can click to view our previous posts, CRISPR Library Screening Applications in Research on Acute Leukemia, CRISPR Screen | Unlocking New Insights into Breast Cancer Targets, CRISPR screen: Opening a New Perspective for Virus Research - Special Guide and so on. In addition, there are many useful CRISPR screening Application posts available for review. We will also launch more CRISPR library related content in the future. Stay tuned!
Based on the above advantages, more and more researchers are choosing CRISPR libraries to screen for new targets and publish high IF articles! However, CRISPR library screening technology is complex, requiring the construction of high-quality libraries, precise cell transduction, and high-throughput sequencing analysis, relying on professional laboratories and personnel.
Ubigene CRISPR-iScreen™platform can provide one-stop services from high-throughput sgRNA library construction to virus packaging, cell infection, drug screening, high-throughput sequencing, and data analysis. Ubigene CRISPR-iScreen™technology utilizing self-developed high efficiency competent cell and exclusive Cell Pool preparation processes, ensures a Cell Pool library coverage of up to 99%.
One-stop CRISPR screening service, screen for new targets as fast as 8 weeks!
400+ CRISPR library in-stock products (transfection-ready plasmids, viruses, cell pools)
Timeline fast as 1 week, various deliverable sizes to choose from.
Inquire now to check the stock availability and price!
Frequently Asked Questions
Q1. What is the difference between RNA sequencing and CRISPR screen?
In summary, RNA-seq measures gene expression levels, while CRISPR screens identify the functional roles of specific genes through genetic perturbation.
1. RNA Sequencing (RNA-seq):
• Definition: Measures gene expression by sequencing RNA molecules.
• Purpose: Identifies which genes are active and their expression levels.
• Outcome: Provides a snapshot of the transcriptome under specific conditions.
• Application: Used to study gene regulation, cellular responses, and differential expression under various conditions.
2. CRISPR Screens:
• Definition: Uses CRISPR technology to systematically knock out or edit genes.
• Purpose: Identifies genes that influence a specific phenotype or response.
• Outcome: Determines which genes are essential or contribute to a particular trait.
• Application: Used to discover essential genes, drug targets, and genetic modifiers of biological processes.
Q2. What are the limitations of the CRISPR screen?
1. Off - target Effects: gRNAs can bind non - target DNA, causing unintended gene disruptions and confusing results.
2. Incomplete Knockout: Mutations may not fully stop gene function, hiding the gene's true role.
3. Cell - type Specificity: Results in one cell type may not apply to others due to different genetic backgrounds.
4. Complex Interactions: It's hard to separate direct and indirect gene effects in genetic networks.
5. Limited to Coding Genes: Traditional screens mainly target coding regions, missing non - coding regulatory elements.
6. Experimental Artifacts: Library quality, transfection efficiency, and selection bias can distort results.
Q3. How does CRISPR screen target identification?
CRISPR screen targets genes with a gRNA library. Each gRNA guides Cas9 to a specific DNA site, where it cuts, creating a double - strand break. Cells repair the break, often causing indels that disrupt gene function. After applying selection pressure (e.g., drug treatment), surviving cells are sequenced. Comparing gRNA abundances in starting and surviving populations helps identify genes crucial for the biological process, such as those involved in cell survival, revealing key genes in biological functions, disease mechanisms, or treatment responses.
References
[1] Joung JK, et al. Genome - scale CRISPR - Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12 (4):828 - 863.
[2] Doench JG, et al. Genome - wide CRISPR - Cas9 knockout screening in human cells. Science. 2016;353 (6304):aaf8729.
[3] Luo B, et al. Genome - wide and pathway - focused CRISPR - Cas9 screens identify mediators of cancer drug resistance. Cell. 2015;161 (6):1247 - 1261.
[4] Reuveni E, et al. Integrating CRISPR screens with multi - omics data to decipher gene regulatory networks. Nat Rev Genet. 2017;18 (7):477 - 490.
[5] Tzelepis K, et al. CRISPR - Cas9 screens in cancer cells to identify novel drug targets and resistance mechanisms. FEBS Lett. 2016;590 (17):2888 - 2901.
[6] Barrera LA, Alvarez-Varela A, Kazane K, et al. CRISPR screens in vivo for tissue-specific cancer drivers. Science. 2016;353 (6307):1160 - 1163.
[7] Tzelepis K, de Stanchina E, D'Arcangelo M, et al. CRISPR-Cas9 screens in cancer cells to identify novel drug targets and resistance mechanisms. FEBS Lett. 2016;590 (17):2888 - 2901.


